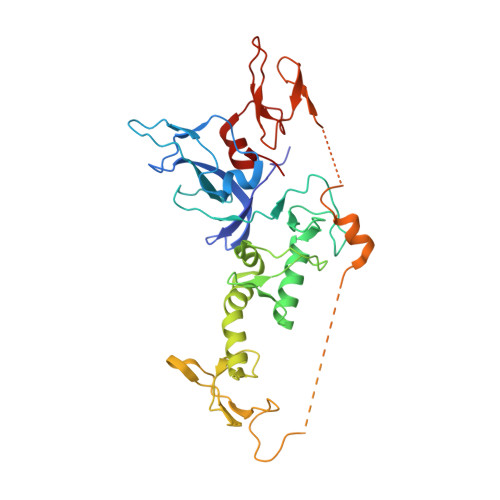
Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Trempe, J.F., Sauve, V., Grenier, K., Seirafi, M., Tang, M.Y., Menade, M., Al-Abdul-Wahid, S., Krett, J., Wong, K., Kozlov, G., Nagar, B., Fon, E.A., Gehring, K.(2013) Science 340: 1451-1455
- PubMed: 23661642
- DOI: https://doi.org/10.1126/science.1237908
- Primary Citation of Related Structures:
4K7D, 4K95 - PubMed Abstract:
Mutations in the PARK2 (parkin) gene are responsible for an autosomal recessive form of Parkinson's disease. The parkin protein is a RING-in-between-RING E3 ubiquitin ligase that exhibits low basal activity. We describe the crystal structure of full-length rat parkin. The structure shows parkin in an autoinhibited state and provides insight into how it is activated. RING0 occludes the ubiquitin acceptor site Cys(431) in RING2, whereas a repressor element of parkin binds RING1 and blocks its E2-binding site. Mutations that disrupted these inhibitory interactions activated parkin both in vitro and in cells. Parkin is neuroprotective, and these findings may provide a structural and mechanistic framework for enhancing parkin activity.
Organizational Affiliation:
McGill Parkinson Program, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montréal, Québec, Canada.