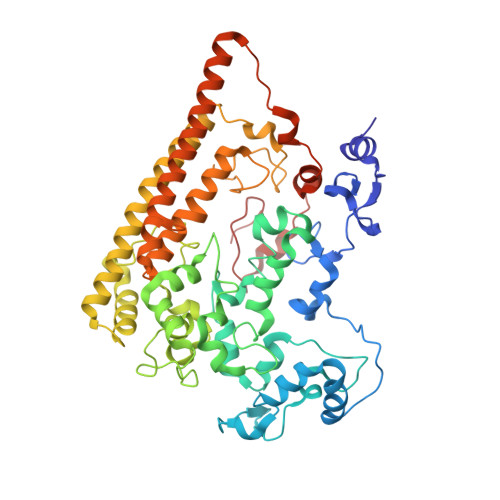
Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
Maalcke, W.J., Dietl, A., Marritt, S.J., Butt, J.N., Jetten, M.S., Keltjens, J.T., Barends, T.R., Kartal, B.(2014) J Biol Chem 289: 1228-1242
- PubMed: 24302732
- DOI: https://doi.org/10.1074/jbc.M113.525147
- Primary Citation of Related Structures:
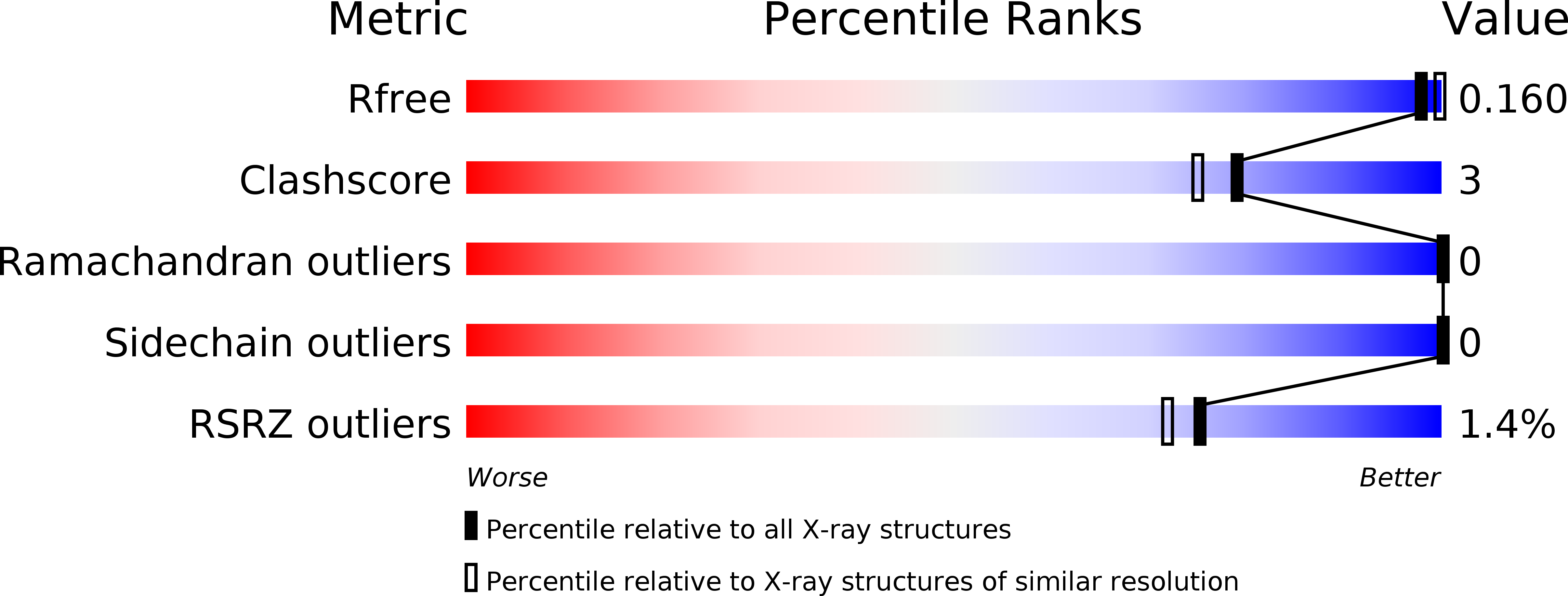
4N4J, 4N4K, 4N4L, 4N4M, 4N4N, 4N4O - PubMed Abstract:
Nitric oxide is an important molecule in all domains of life with significant biological functions in both pro- and eukaryotes. Anaerobic ammonium-oxidizing (anammox) bacteria that contribute substantially to the release of fixed nitrogen into the atmosphere use the oxidizing power of NO to activate inert ammonium into hydrazine (N2H4). Here, we describe an enzyme from the anammox bacterium Kuenenia stuttgartiensis that uses a novel pathway to make NO from hydroxylamine. This new enzyme is related to octaheme hydroxylamine oxidoreductase, a key protein in aerobic ammonium-oxidizing bacteria. By a multiphasic approach including the determination of the crystal structure of the K. stuttgartiensis enzyme at 1.8 Å resolution and refinement and reassessment of the hydroxylamine oxidoreductase structure from Nitrosomonas europaea, both in the presence and absence of their substrates, we propose a model for NO formation by the K. stuttgartiensis enzyme. Our results expand the understanding of the functions that the widespread family of octaheme proteins have.
Organizational Affiliation:
From the Department of Microbiology, Institute for Water and Wetland Research, Radboud University Nijmegen, 6525AJ Nijmegen, The Netherlands.