Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chen, L., Wang, Z.G., Aleshin, A.E., Chen, F., Chen, J., Jiang, F., Alitongbieke, G., Zeng, Z., Ma, Y., Huang, M., Zhou, H., Cadwell, G., Zheng, J.F., Huang, P.Q., Liddington, R.C., Zhang, X.K., Su, Y.(2014) Chem Biol 21: 596-607
- PubMed: 24704507
- DOI: https://doi.org/10.1016/j.chembiol.2014.02.017
- Primary Citation of Related Structures:
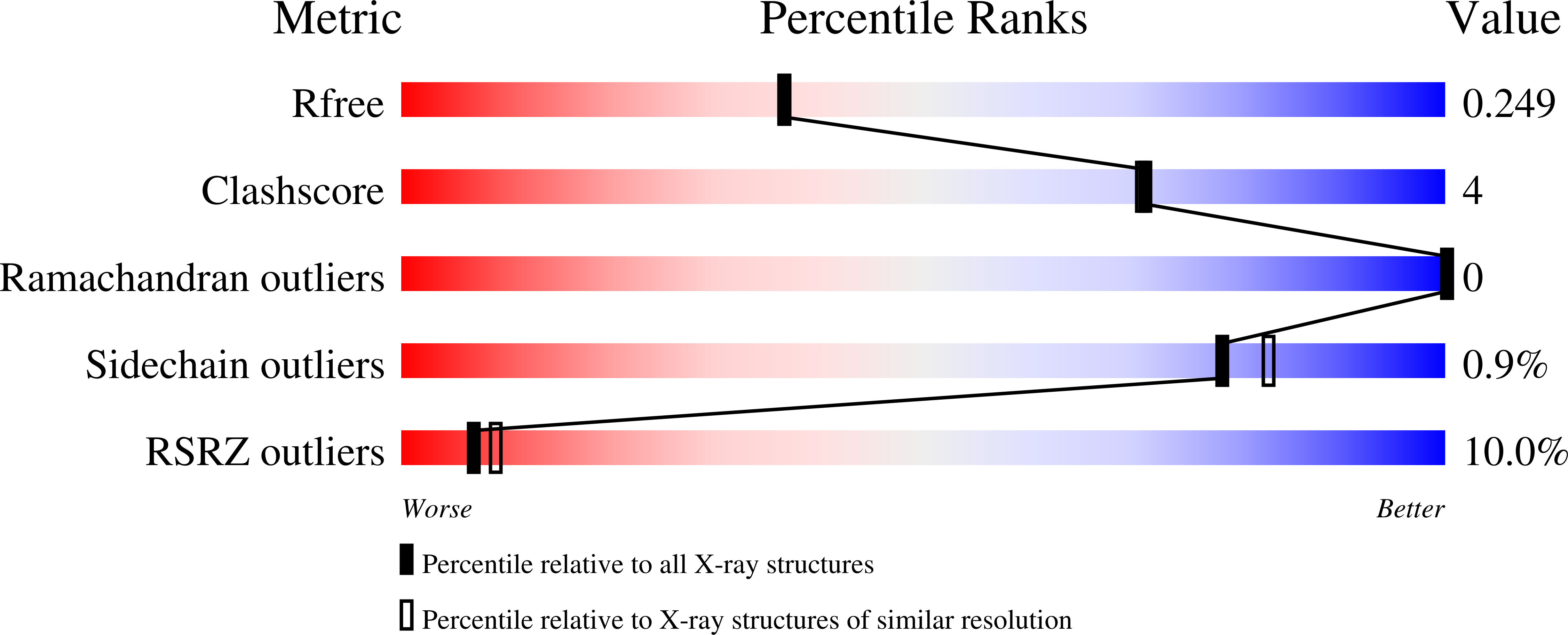
4N5G, 4N8R - PubMed Abstract:
Retinoid X receptor-alpha (RXRα), an intriguing and unique drug target, can serve as an intracellular target mediating the anticancer effects of certain nonsteroidal anti-inflammatory drugs (NSAIDs), including sulindac. We report the synthesis and characterization of two sulindac analogs, K-8008 and K-8012, which exert improved anticancer activities over sulindac in a RXRα-dependent manner. The analogs inhibit the interaction of the N-terminally truncated RXRα (tRXRα) with the p85α subunit of PI3K, leading to suppression of AKT activation and induction of apoptosis. Crystal structures of the RXRα ligand-binding domain (LBD) with K-8008 or K-8012 reveal that both compounds bind to tetrameric RXRα LBD at a site different from the classical ligand-binding pocket. Thus, these results identify K-8008 and K-8012 as tRXRα modulators and define a binding mechanism for regulating the nongenomic action of tRXRα.
Organizational Affiliation:
School of Pharmaceutical Sciences, Xiamen University, Xiamen 361102, China; Sanford-Burnham Medical Research Institute, 10901 North Torrey Pines Road, La Jolla, CA 92037, USA.