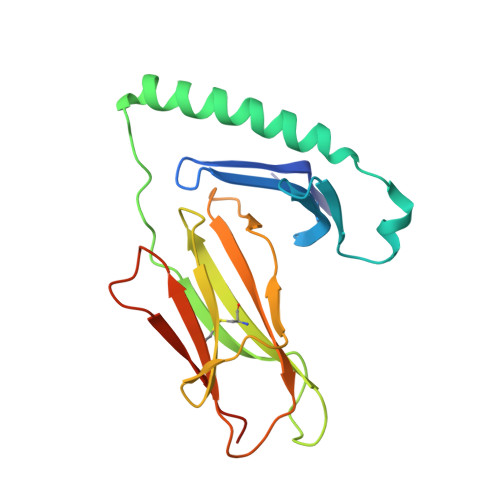
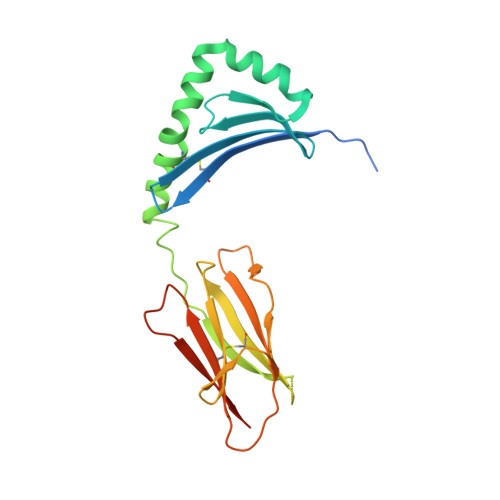
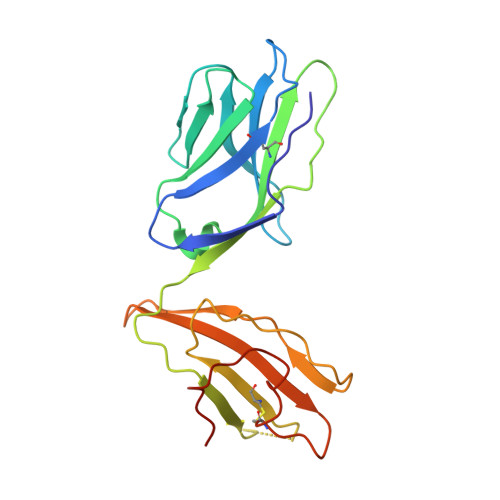
T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease.
Petersen, J., Montserrat, V., Mujico, J.R., Loh, K.L., Beringer, D.X., van Lummel, M., Thompson, A., Mearin, M.L., Schweizer, J., Kooy-Winkelaar, Y., van Bergen, J., Drijfhout, J.W., Kan, W.T., La Gruta, N.L., Anderson, R.P., Reid, H.H., Koning, F., Rossjohn, J.(2014) Nat Struct Mol Biol 21: 480-488
- PubMed: 24777060
- DOI: https://doi.org/10.1038/nsmb.2817
- Primary Citation of Related Structures:
4OZF, 4OZG, 4OZH, 4OZI - PubMed Abstract:
Celiac disease is a T cell-mediated disease induced by dietary gluten, a component of which is gliadin. 95% of individuals with celiac disease carry the HLA (human leukocyte antigen)-DQ2 locus. Here we determined the T-cell receptor (TCR) usage and fine specificity of patient-derived T-cell clones specific for two epitopes from wheat gliadin, DQ2.5-glia-α1a and DQ2.5-glia-α2. We determined the ternary structures of four distinct biased TCRs specific for those epitopes. All three TCRs specific for DQ2.5-glia-α2 docked centrally above HLA-DQ2, which together with mutagenesis and affinity measurements provided a basis for the biased TCR usage. A non-germline encoded arginine residue within the CDR3β loop acted as the lynchpin within this common docking footprint. Although the TCRs specific for DQ2.5-glia-α1a and DQ2.5-glia-α2 docked similarly, their interactions with the respective gliadin determinants differed markedly, thereby providing a basis for epitope specificity.
Organizational Affiliation:
1] Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria, Australia. [2].