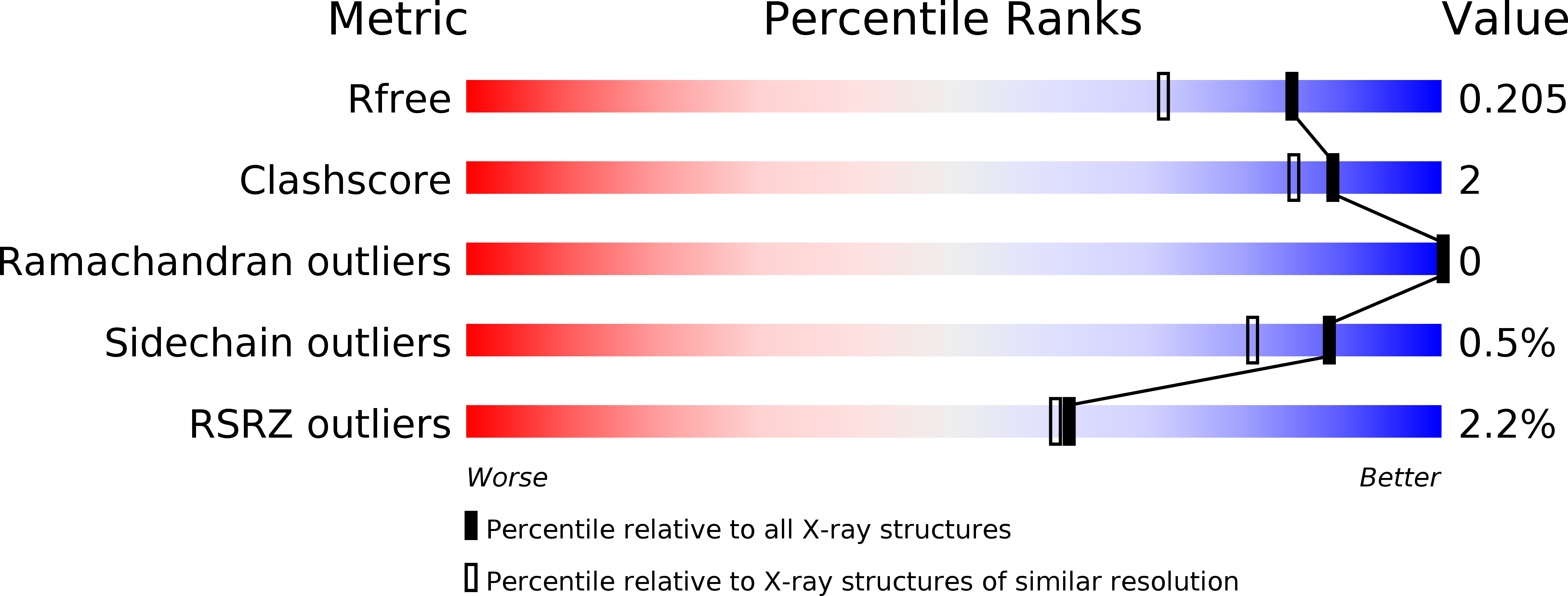
Structure, biochemistry, and inhibition of essential 4'-phosphopantetheinyl transferases from two species of mycobacteria.
Vickery, C.R., Kosa, N.M., Casavant, E.P., Duan, S., Noel, J.P., Burkart, M.D.(2014) ACS Chem Biol 9: 1939-1944
- PubMed: 24963544
- DOI: https://doi.org/10.1021/cb500263p
- Primary Citation of Related Structures:
4QJK, 4QJL - PubMed Abstract:
4'-Phosphopantetheinyl transferases (PPTase) post-translationally modify carrier proteins with a phosphopantetheine moiety, an essential reaction in all three domains of life. In the bacterial genus Mycobacteria, the Sfp-type PPTase activates pathways necessary for the biosynthesis of cell wall components and small molecule virulence factors. We solved the X-ray crystal structures and biochemically characterized the Sfp-type PPTases from two of the most prevalent Mycobacterial pathogens, PptT of M. tuberculosis and MuPPT of M. ulcerans. Structural analyses reveal significant differences in cofactor binding and active site composition when compared to previously characterized Sfp-type PPTases. Functional analyses including the efficacy of Sfp-type PPTase-specific inhibitors also suggest that the Mycobacterial Sfp-type PPTases can serve as therapeutic targets against Mycobacterial infections.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego , 9500 Gilman Drive, La Jolla, California 92093-0358, United States.