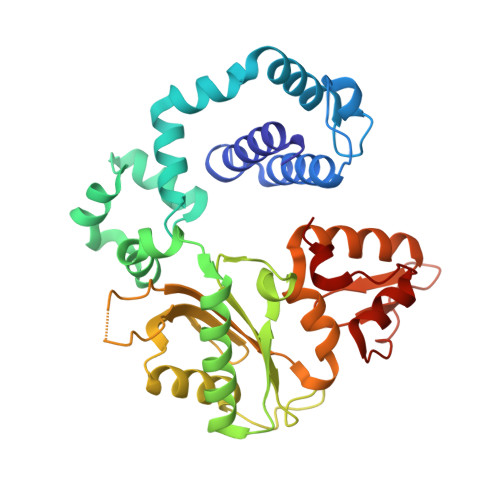
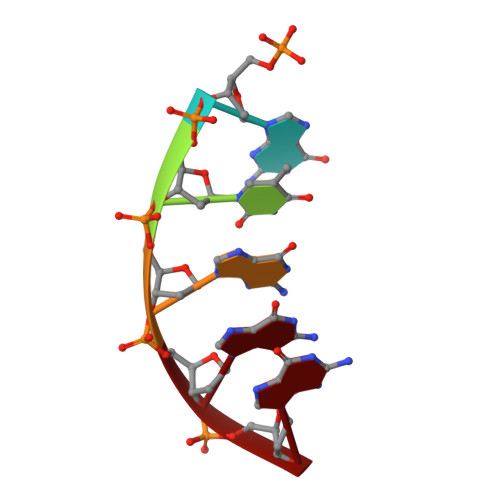
Structural Basis for the Inefficient Nucleotide Incorporation Opposite Cisplatin-DNA Lesion by Human DNA Polymerase beta.
Koag, M.C., Lai, L., Lee, S.(2014) J Biol Chem 289: 31341-31348
- PubMed: 25237188
- DOI: https://doi.org/10.1074/jbc.M114.605451
- Primary Citation of Related Structures:
4TUP, 4TUQ, 4TUR, 4TUS - PubMed Abstract:
Human DNA polymerase β (polβ) has been suggested to play a role in cisplatin resistance, especially in polβ-overexpressing cancer cells. Polβ has been shown to accurately albeit slowly bypass the cisplatin-1,2-d(GpG) (Pt-GG) intramolecular cross-link in vitro. Currently, the structural basis for the inefficient Pt-GG bypass mechanism of polβ is unknown. To gain structural insights into the mechanism, we determined two ternary structures of polβ incorporating dCTP opposite the templating Pt-GG lesion in the presence of the active site Mg(2+) or Mn(2+). The Mg(2+)-bound structure shows that the bulky Pt-GG adduct is accommodated in the polβ active site without any steric hindrance. In addition, both guanines of the Pt-GG lesion form Watson-Crick base pairing with the primer terminus dC and the incoming dCTP, providing the structural basis for the accurate bypass of the Pt-GG adduct by polβ. The Mn(2+)-bound structure shows that polβ adopts a catalytically suboptimal semiclosed conformation during the insertion of dCTP opposite the templating Pt-GG, explaining the inefficient replication across the Pt-GG lesion by polβ. Overall, our studies provide the first structural insights into the mechanism of the potential polβ-mediated cisplatin resistance.
Organizational Affiliation:
From the Division of Medicinal Chemistry, College of Pharmacy, The University of Texas, Austin, Texas 78712.