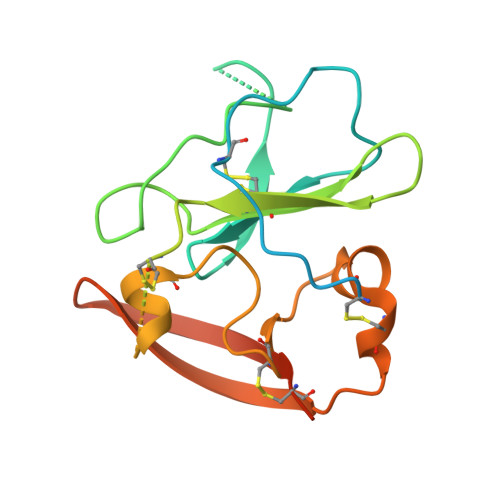
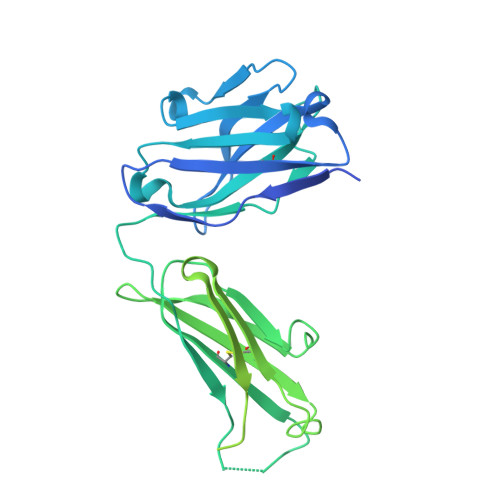
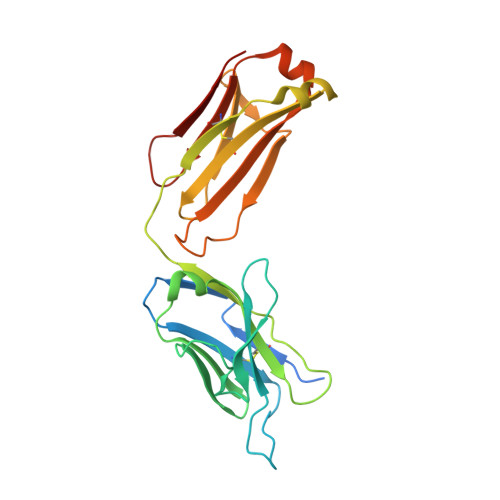
Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2.
Khan, A.G., Whidby, J., Miller, M.T., Scarborough, H., Zatorski, A.V., Cygan, A., Price, A.A., Yost, S.A., Bohannon, C.D., Jacob, J., Grakoui, A., Marcotrigiano, J.(2014) Nature 509: 381-384
- PubMed: 24553139
- DOI: https://doi.org/10.1038/nature13117
- Primary Citation of Related Structures:
4WEB - PubMed Abstract:
Hepatitis C virus (HCV) is a significant public health concern with approximately 160 million people infected worldwide. HCV infection often results in chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. No vaccine is available and current therapies are effective against some, but not all, genotypes. HCV is an enveloped virus with two surface glycoproteins (E1 and E2). E2 binds to the host cell through interactions with scavenger receptor class B type I (SR-BI) and CD81, and serves as a target for neutralizing antibodies. Little is known about the molecular mechanism that mediates cell entry and membrane fusion, although E2 is predicted to be a class II viral fusion protein. Here we describe the structure of the E2 core domain in complex with an antigen-binding fragment (Fab) at 2.4 Å resolution. The E2 core has a compact, globular domain structure, consisting mostly of β-strands and random coil with two small α-helices. The strands are arranged in two, perpendicular sheets (A and B), which are held together by an extensive hydrophobic core and disulphide bonds. Sheet A has an IgG-like fold that is commonly found in viral and cellular proteins, whereas sheet B represents a novel fold. Solution-based studies demonstrate that the full-length E2 ectodomain has a similar globular architecture and does not undergo significant conformational or oligomeric rearrangements on exposure to low pH. Thus, the IgG-like fold is the only feature that E2 shares with class II membrane fusion proteins. These results provide unprecedented insights into HCV entry and will assist in developing an HCV vaccine and new inhibitors.
Organizational Affiliation:
Center for Advanced Biotechnology and Medicine, Department of Chemistry and Chemical Biology, Rutgers University, 679 Hoes Lane West, Piscataway, New Jersey 08854, USA.