Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
Hubrich, F., Juneja, P., Mueller, M., Diederichs, K., Welte, W., Andexer, J.N.(2015) J Am Chem Soc 137: 11032
- PubMed: 26247872
- DOI: https://doi.org/10.1021/jacs.5b05559
- Primary Citation of Related Structures:
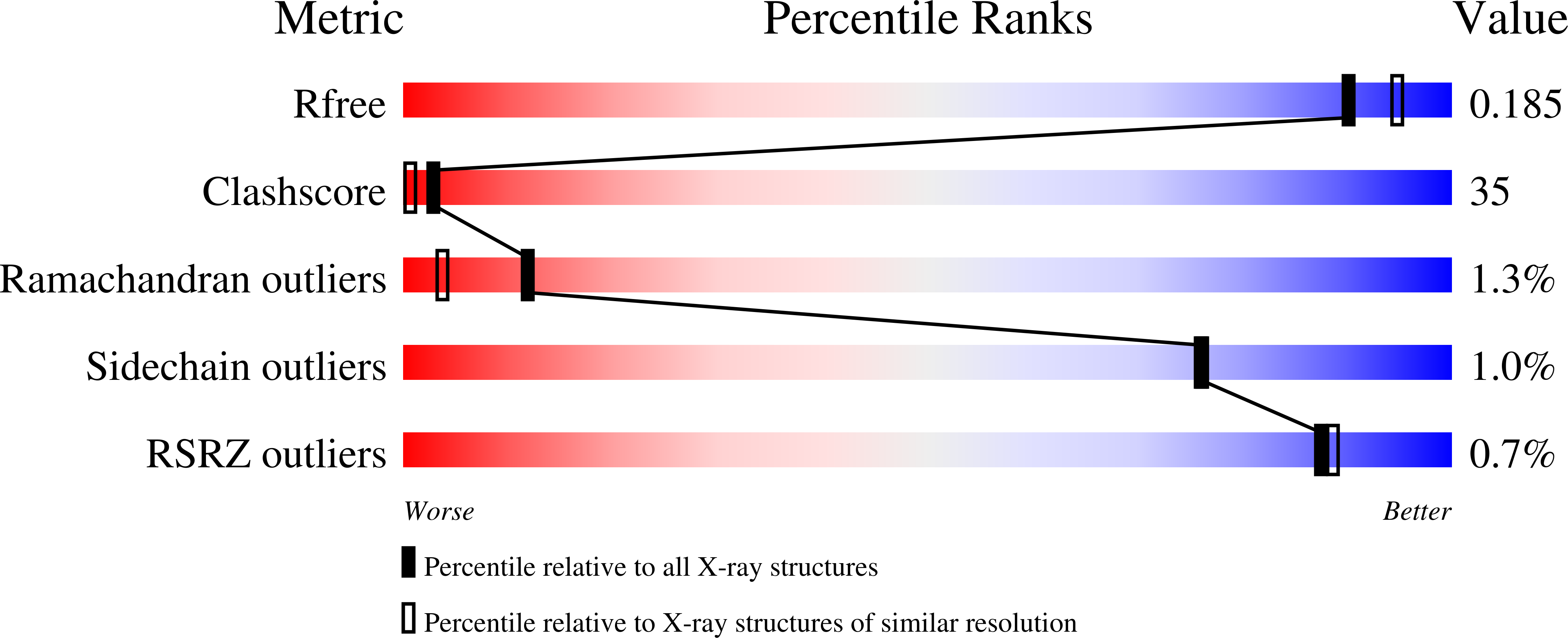
5A3K, 5AG3 - PubMed Abstract:
Chorismatases are a class of chorismate-converting enzymes involved in the biosynthetic pathways of different natural products, many of them with interesting pharmaceutical characteristics. So far, three subfamilies of chorismatases are described that convert chorismate into different (dihydro-)benzoate derivatives (CH-FkbO, CH-Hyg5, and CH-XanB2). Until now, the detailed enzyme mechanism and the molecular basis for the different reaction products were unknown. Here we show that the CH-FkbO and CH-Hyg5 subfamilies share the same protein fold, but employ fundamentally different reaction mechanisms. While the FkbO reaction is a typical hydrolysis, the Hyg5 reaction proceeds intramolecularly, most likely via an arene oxide intermediate. Two nonconserved active site residues were identified that are responsible for the different reaction mechanisms in CH-FkbO and CH-Hyg5. Further, we propose an additional amino acid residue to be responsible for the discrimination of the CH-XanB2 subfamily, which catalyzes the formation of two different hydroxybenzoate regioisomers, likely in a single active site. A multiple sequence alignment shows that these three crucial amino acid positions are located in conserved motifs and can therefore be used to assign unknown chorismatases to the corresponding subfamily.
Organizational Affiliation:
Institute of Pharmaceutical Sciences, University of Freiburg , Albertstr. 25, 79104 Freiburg, Germany.