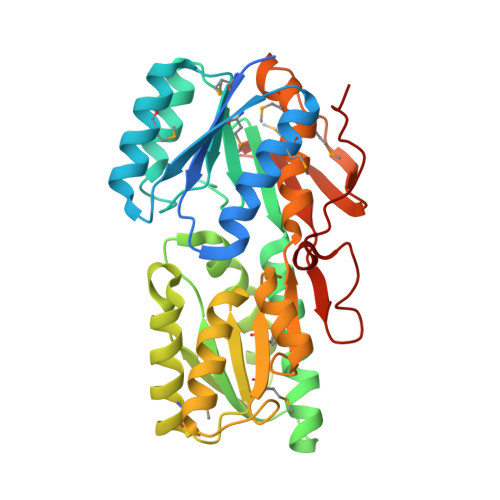
Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Yadava, U., Vetting, M.W., Al Obaidi, N., Carter, M.S., Gerlt, J.A., Almo, S.C.(2016) Acta Crystallogr F Struct Biol Commun 72: 467-472
- PubMed: 27303900
- DOI: https://doi.org/10.1107/S2053230X16007500
- Primary Citation of Related Structures:
4Y9T, 5BR1 - PubMed Abstract:
The uptake of exogenous solutes by prokaryotes is mediated by transport systems embedded in the plasma membrane. In many cases, a solute-binding protein (SBP) is utilized to bind ligands with high affinity and deliver them to the membrane-bound components responsible for translocation into the cytoplasm. In the present study, Avi_5305, an Agrobacterium vitis SBP belonging to Pfam13407, was screened by differential scanning fluorimetry (DSF) and found to be stabilized by D-glucosamine and D-galactosamine. Avi_5305 is the first protein from Pfam13407 shown to be specific for amino sugars, and co-crystallization resulted in structures of Avi_5305 bound to D-glucosamine and D-galactosamine. Typical of Pfam13407, Avi_5305 consists of two α/β domains linked through a hinge region, with the ligand-binding site located in a cleft between the two domains. Comparisons with Escherichia coli ribose-binding protein suggest that a cation-π interaction with Tyr168 provides the specificity for D-glucosamine/D-galactosamine over D-glucose/D-galactose.
Organizational Affiliation:
Department of Physics, DDU Gorakhpur University, Gorakhpur 273 009, India.