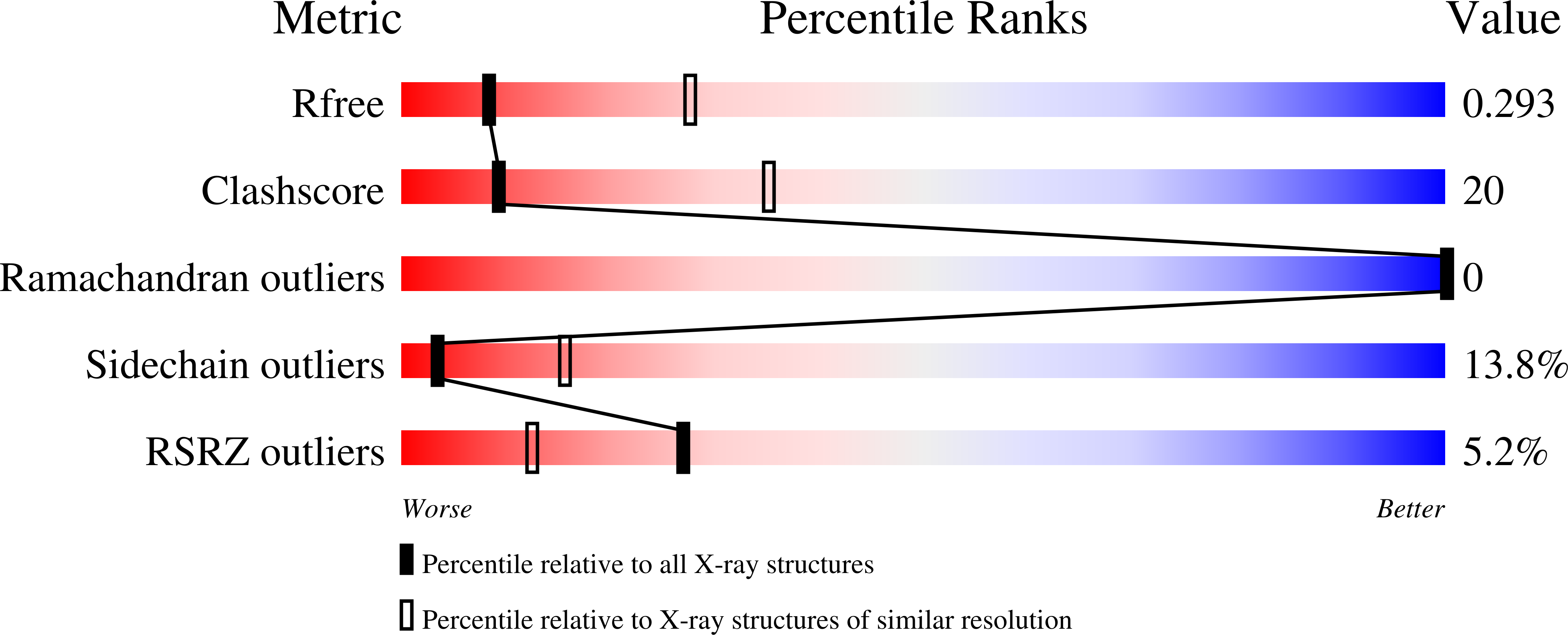
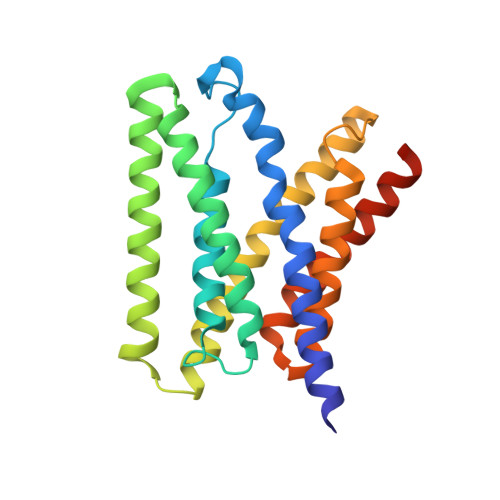
Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Tao, Y., Cheung, L.S., Li, S., Eom, J.S., Chen, L.Q., Xu, Y., Perry, K., Frommer, W.B., Feng, L.(2015) Nature 527: 259-263
- PubMed: 26479032
- DOI: https://doi.org/10.1038/nature15391
- Primary Citation of Related Structures:
5CTG, 5CTH - PubMed Abstract:
Eukaryotes rely on efficient distribution of energy and carbon skeletons between organs in the form of sugars. Glucose in animals and sucrose in plants serve as the dominant distribution forms. Cellular sugar uptake and release require vesicular and/or plasma membrane transport proteins. Humans and plants use proteins from three superfamilies for sugar translocation: the major facilitator superfamily (MFS), the sodium solute symporter family (SSF; only in the animal kingdom), and SWEETs. SWEETs carry mono- and disaccharides across vacuolar or plasma membranes. Plant SWEETs play key roles in sugar translocation between compartments, cells, and organs, notably in nectar secretion, phloem loading for long distance translocation, pollen nutrition, and seed filling. Plant SWEETs cause pathogen susceptibility possibly by sugar leakage from infected cells. The vacuolar Arabidopsis thaliana AtSWEET2 sequesters sugars in root vacuoles; loss-of-function mutants show increased susceptibility to Pythium infection. Here we show that its orthologue, the vacuolar glucose transporter OsSWEET2b from rice (Oryza sativa), consists of an asymmetrical pair of triple-helix bundles, connected by an inversion linker transmembrane helix (TM4) to create the translocation pathway. Structural and biochemical analyses show OsSWEET2b in an apparent inward (cytosolic) open state forming homomeric trimers. TM4 tightly interacts with the first triple-helix bundle within a protomer and mediates key contacts among protomers. Structure-guided mutagenesis of the close paralogue SWEET1 from Arabidopsis identified key residues in substrate translocation and protomer crosstalk. Insights into the structure-function relationship of SWEETs are valuable for understanding the transport mechanism of eukaryotic SWEETs and may be useful for engineering sugar flux.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, 279 Campus Drive, Stanford University School of Medicine, Stanford, CA 94305, USA.