Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Miyanaga, A., Iwasawa, S., Shinohara, Y., Kudo, F., Eguchi, T.(2016) Proc Natl Acad Sci U S A 113: 1802-1807
- PubMed: 26831085
- DOI: https://doi.org/10.1073/pnas.1520042113
- Primary Citation of Related Structures:
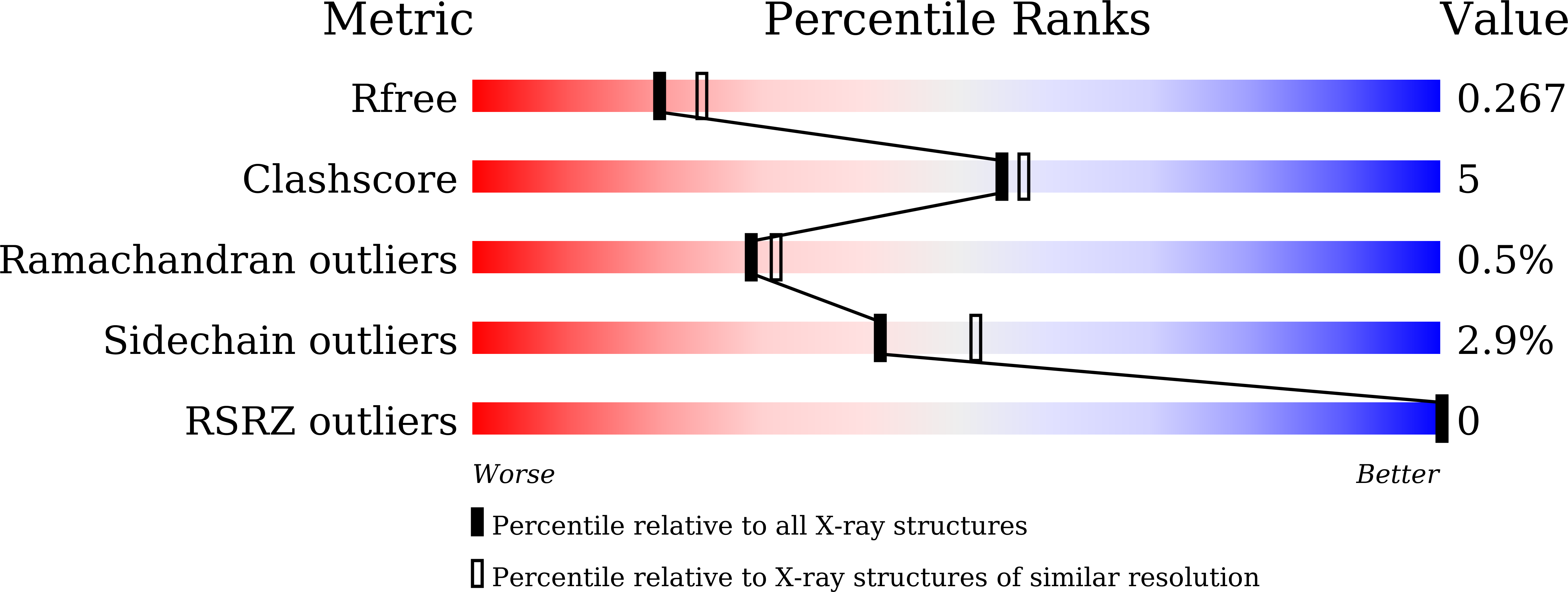
5CZC, 5CZD - PubMed Abstract:
Acyltransferases (ATs) are key determinants of building block specificity in polyketide biosynthesis. Despite the importance of protein-protein interactions between AT and acyl carrier protein (ACP) during the acyltransfer reaction, the mechanism of ACP recognition by AT is not understood in detail. Herein, we report the crystal structure of AT VinK, which transfers a dipeptide group between two ACPs, VinL and VinP1LdACP, in vicenistatin biosynthesis. The isolated VinK structure showed a unique substrate-binding pocket for the dipeptide group linked to ACP. To gain greater insight into the mechanism of ACP recognition, we attempted to crystallize the VinK-ACP complexes. Because transient enzyme-ACP complexes are difficult to crystallize, we developed a covalent cross-linking strategy using a bifunctional maleimide reagent to trap the VinK-ACP complexes, allowing the determination of the crystal structure of the VinK-VinL complex. In the complex structure, Arg-153, Met-206, and Arg-299 of VinK interact with the negatively charged helix II region of VinL. The VinK-VinL complex structure allows, to our knowledge, the first visualization of the interaction between AT and ACP and provides detailed mechanistic insights into ACP recognition by AT.
Organizational Affiliation:
Department of Chemistry, Tokyo Institute of Technology, O-okayama, Meguro-ku, Tokyo 152-8551, Japan; [email protected] [email protected].