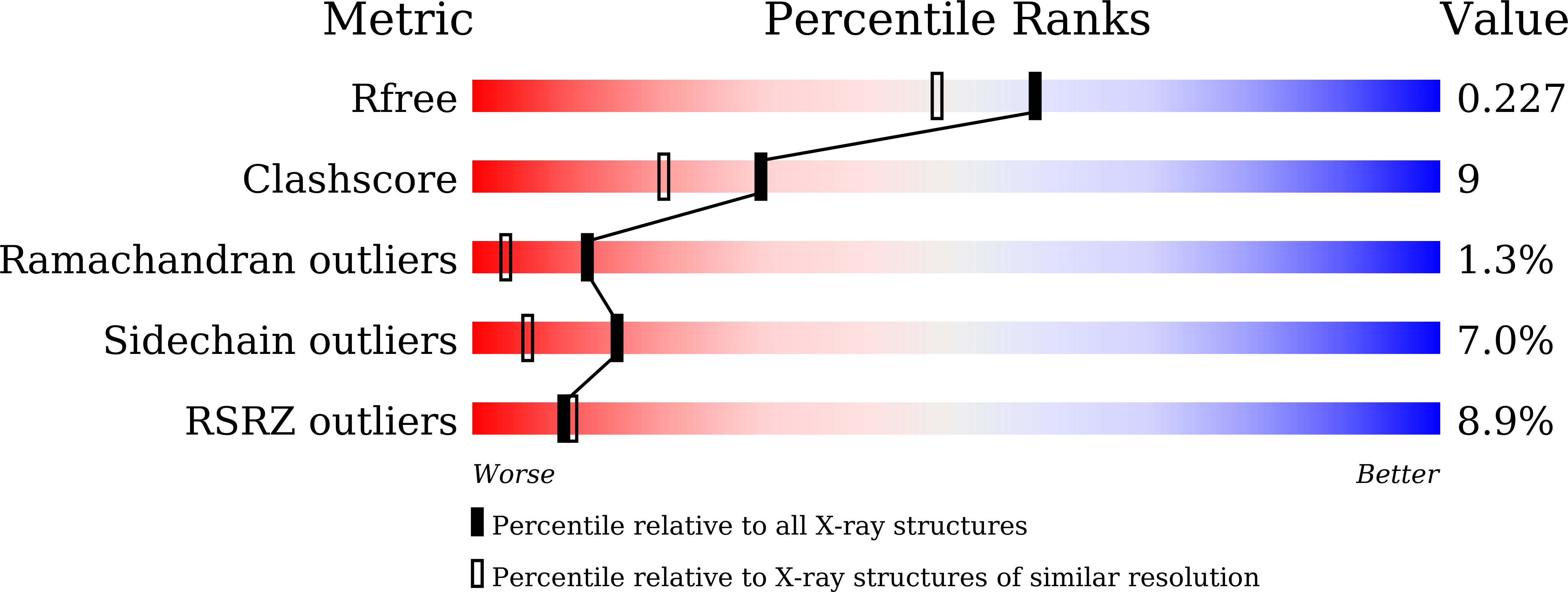
Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Liu, M.X., Chen, C.C., Chen, L., Xiao, X.S., Zheng, Y.Y., Huang, J.W., Liu, W.d., Ko, T.P., Cheng, Y.S., Feng, X.X., Oldfield, E., Guo, R.T., Ma, Y.H.(2016) Angew Chem Int Ed Engl 55: 4721-4724
- PubMed: 26922900
- DOI: https://doi.org/10.1002/anie.201600656
- Primary Citation of Related Structures:
5HC6, 5HC7, 5HC8 - PubMed Abstract:
We report the first X-ray structure of the unique "head-to-middle" monoterpene synthase, lavandulyl diphosphate synthase (LPPS). LPPS catalyzes the condensation of two molecules of dimethylallyl diphosphate (DMAPP) to form lavandulyl diphosphate, a precursor to the fragrance lavandulol. The structure is similar to that of the bacterial cis-prenyl synthase, undecaprenyl diphosphate synthase (UPPS), and contains an allylic site (S1) in which DMAPP ionizes and a second site (S2) which houses the DMAPP nucleophile. Both S-thiolo-dimethylallyl diphosphate and S-thiolo-isopentenyl diphosphate bind intact to S2, but are cleaved to (thio)diphosphate, in S1. His78 (Asn in UPPS) is essential for catalysis and is proposed to facilitate diphosphate release in S1, while the P1 phosphate in S2 abstracts a proton from the lavandulyl carbocation to form the LPP product. The results are of interest since they provide the first structure and structure-based mechanism of this unusual prenyl synthase.
Organizational Affiliation:
College of Biotechnology, Tianjin University of Science and Technology, Tianjin 300457, China.