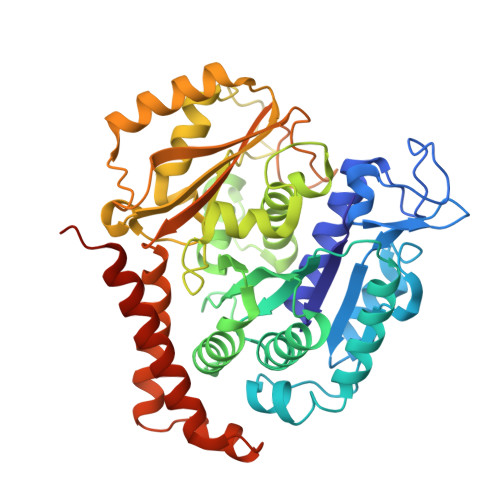
Structural Basis of Microtubule Destabilization by Potent Auristatin Anti-Mitotics.
Waight, A.B., Bargsten, K., Doronina, S., Steinmetz, M.O., Sussman, D., Prota, A.E.(2016) PLoS One 11: e0160890-e0160890
- PubMed: 27518442
- DOI: https://doi.org/10.1371/journal.pone.0160890
- Primary Citation of Related Structures:
5IYZ, 5J2T, 5J2U - PubMed Abstract:
The auristatin class of microtubule destabilizers are highly potent cytotoxic agents against several cancer cell types when delivered as antibody drug conjugates. Here we describe the high resolution structures of tubulin in complex with both monomethyl auristatin E and F and unambiguously define the trans-configuration of both ligands at the Val-Dil amide bond in their tubulin bound state. Moreover, we illustrate how peptidic vinca-site agents carrying terminal carboxylate residues may exploit an observed extended hydrogen bond network with the M-loop Arg278 to greatly improve the affinity of the corresponding analogs and to maintain the M-loop in an incompatible conformation for productive lateral tubulin-tubulin contacts in microtubules. Our results highlight a potential, previously undescribed molecular mechanism by which peptidic vinca-site agents maintain unparalleled potency as microtubule-destabilizing agents.
Organizational Affiliation:
Department of Protein Sciences, Seattle Genetics, Inc., Bothell, WA, United States of America.