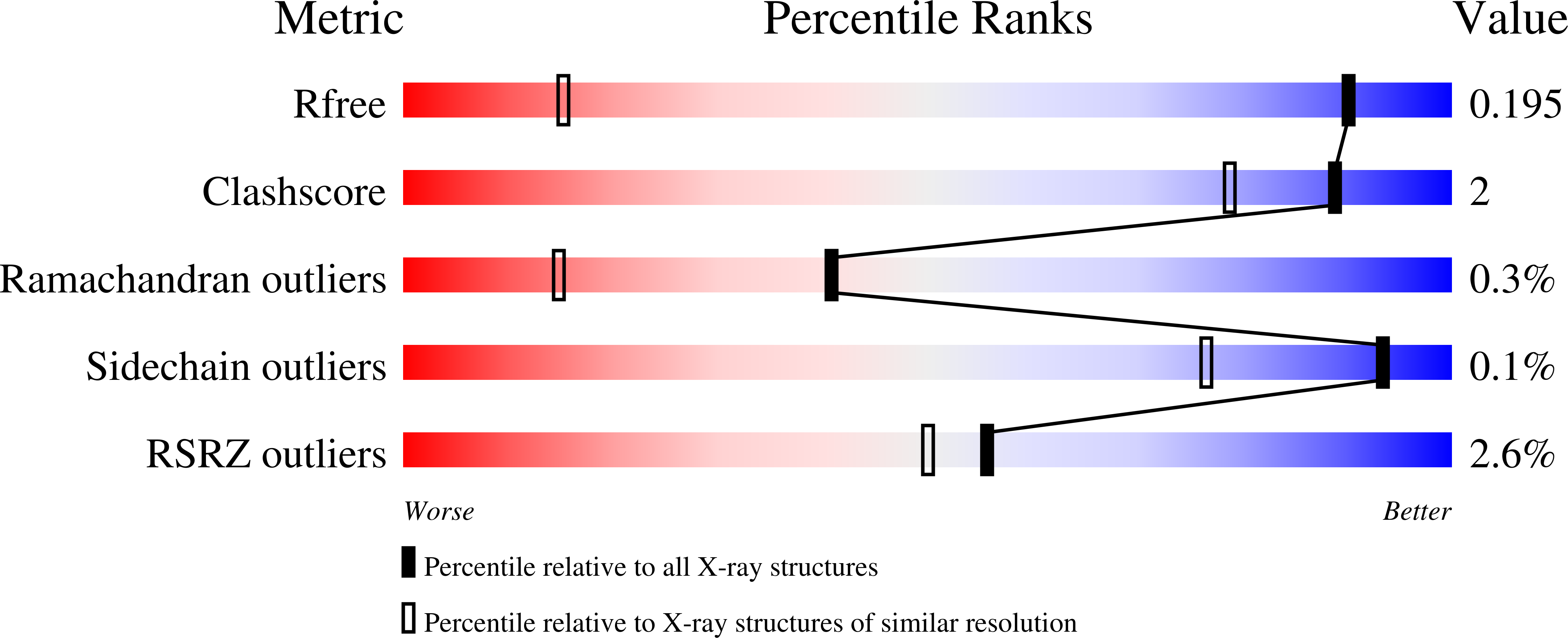
Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Zhang, Z., Smart, T.J., Choi, H., Hardy, F., Lohans, C.T., Abboud, M.I., Richardson, M.S.W., Paton, R.S., McDonough, M.A., Schofield, C.J.(2017) Proc Natl Acad Sci U S A 114: 4667-4672
- PubMed: 28420789
- DOI: https://doi.org/10.1073/pnas.1617760114
- Primary Citation of Related Structures:
5LSQ, 5LUN, 5MOF - PubMed Abstract:
Ethylene is important in industry and biological signaling. In plants, ethylene is produced by oxidation of 1-aminocyclopropane-1-carboxylic acid, as catalyzed by 1-aminocyclopropane-1-carboxylic acid oxidase. Bacteria catalyze ethylene production, but via the four-electron oxidation of 2-oxoglutarate to give ethylene in an arginine-dependent reaction. Crystallographic and biochemical studies on the Pseudomonas syringae ethylene-forming enzyme reveal a branched mechanism. In one branch, an apparently typical 2-oxoglutarate oxygenase reaction to give succinate, carbon dioxide, and sometimes pyrroline-5-carboxylate occurs. Alternatively, Grob-type oxidative fragmentation of a 2-oxoglutarate-derived intermediate occurs to give ethylene and carbon dioxide. Crystallographic and quantum chemical studies reveal that fragmentation to give ethylene is promoted by binding of l-arginine in a nonoxidized conformation and of 2-oxoglutarate in an unprecedented high-energy conformation that favors ethylene, relative to succinate formation.
Organizational Affiliation:
Department of Chemistry, University of Oxford, Oxford, OX1 3TA, United Kingdom.