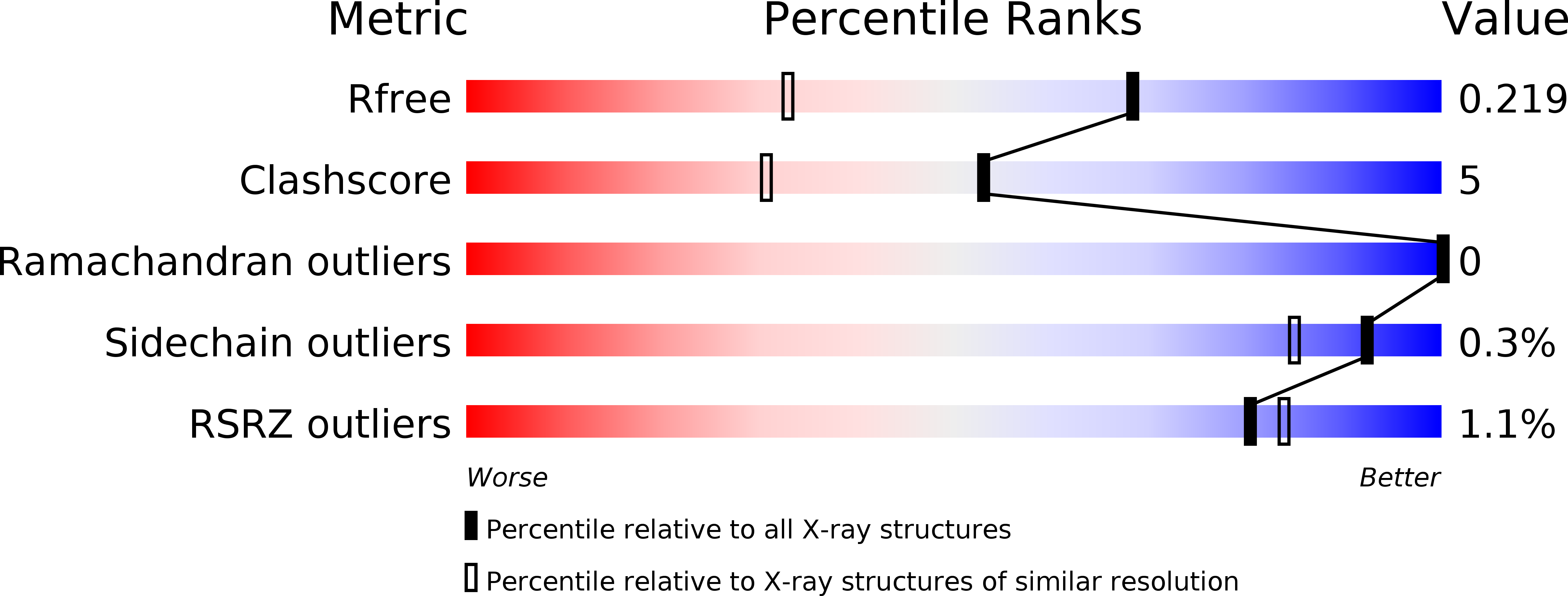
Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
He, R., Di Bonaventura, I., Visini, R., Gan, B.H., Fu, Y., Probst, D., Luscher, A., Kohler, T., van Delden, C., Stocker, A., Hong, W., Darbre, T., Reymond, J.L.(2017) Chem Sci 8: 7464-7475
- PubMed: 29163899
- DOI: https://doi.org/10.1039/c7sc01599b
- Primary Citation of Related Structures:
5NES, 5NEY, 5NF0 - PubMed Abstract:
Here we report a new family of cyclic antimicrobial peptides (CAMPs) targeting MDR strains of Pseudomonas aeruginosa . These CAMPs are cyclized via a xylene double thioether bridge connecting two cysteines placed at the ends of a linear amphiphilic alternating d,l-sequence composed of lysines and tryptophans. Investigations by transmission electron microscopy (TEM), dynamic light scattering and atomic force microscopy (AFM) suggest that these peptide macrocycles interact with the membrane to form lipid-peptide aggregates. Amphiphilic conformations compatible with membrane disruption are observed in high resolution X-ray crystal structures of fucosylated derivatives in complex with lectin LecB. The potential for optimization is highlighted by N -methylation of backbone amides leading to derivatives with similar antimicrobial activity but lower hemolysis.
Organizational Affiliation:
Department of Chemistry and Biochemistry , University of Bern , Freiestrasse 3 , 3012 Bern , Switzerland . Email: [email protected].