Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Hochscherf, J., Lindenblatt, D., Witulski, B., Birus, R., Aichele, D., Marminon, C., Bouaziz, Z., Le Borgne, M., Jose, J., Niefind, K.(2017) Pharmaceuticals (Basel) 10
- PubMed: 29236079
- DOI: https://doi.org/10.3390/ph10040098
- Primary Citation of Related Structures:
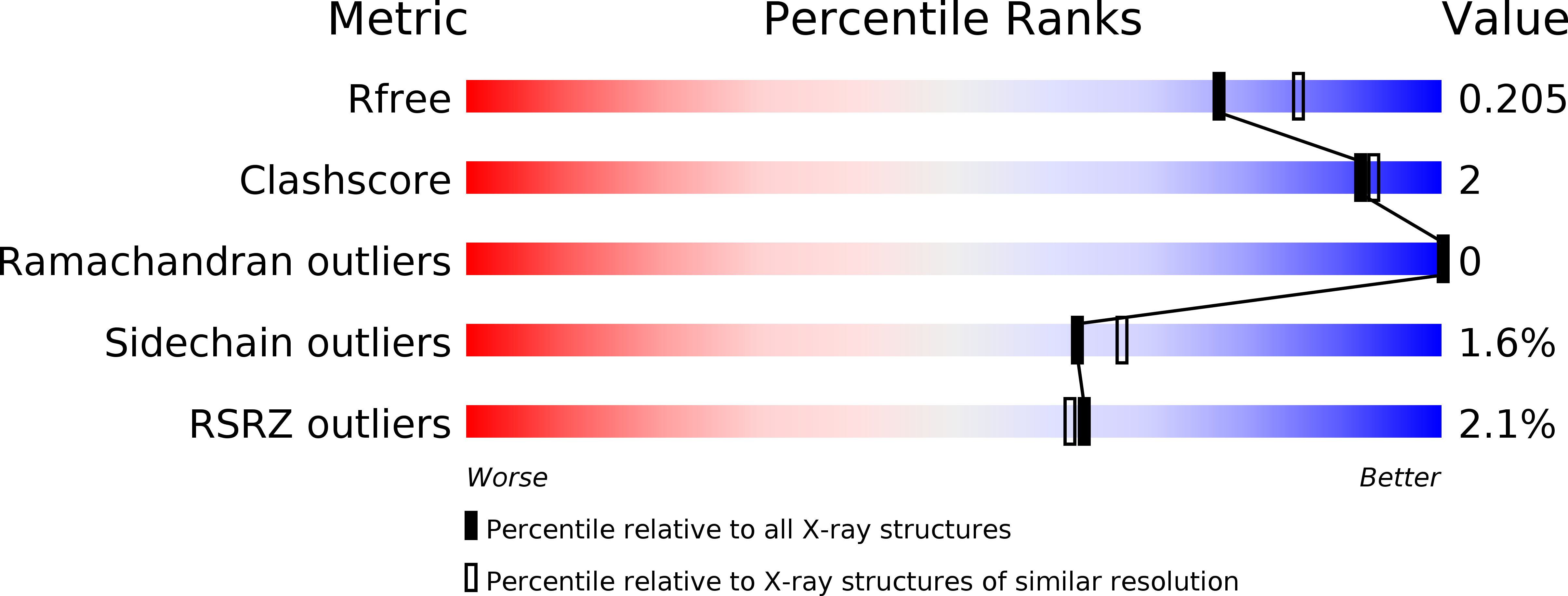
5OMY, 5ONI, 5OOI - PubMed Abstract:
Protein kinase CK2, a member of the eukaryotic protein kinase superfamily, is associated with cancer and other human pathologies and thus an attractive drug target. The indeno[1,2- b ]indole scaffold is a novel lead structure to develop ATP-competitive CK2 inhibitors. Some indeno[1,2- b ]indole-based CK2 inhibitors additionally obstruct ABCG2, an ABC half transporter overexpressed in breast cancer and co-responsible for drug efflux and resistance. Comprehensive derivatization studies revealed substitutions of the indeno[1,2- b ]indole framework that boost either the CK2 or the ABCG2 selectivity or even support the dual inhibition potential. The best indeno[1,2- b ]indole-based CK2 inhibitor described yet (IC 50 = 25 nM) is 5-isopropyl-4-(3-methylbut-2-enyl-oxy)-5,6,7,8-tetrahydroindeno[1,2- b ]indole-9,10-dione ( 4p ). Herein, we demonstrate the membrane permeability of 4p and describe co-crystal structures of 4p with CK2α and CK2α', the paralogs of human CK2 catalytic subunit. As expected, 4p occupies the narrow, hydrophobic ATP site of CK2α/CK2α', but surprisingly with a unique orientation: its hydrophobic substituents point towards the solvent while its two oxo groups are hydro-gen-bonded to a hidden water molecule. An equivalent water molecule was found in many CK2α structures, but never as a critical mediator of ligand binding. This unexpected binding mode is independent of the interdomain hinge/helix αD region conformation and of the salt content in the crystallization medium.
Organizational Affiliation:
Department für Chemie, Institut für Biochemie, Universität zu Köln, Zülpicher Straße 47, D-50674 Köln, Germany. [email protected].