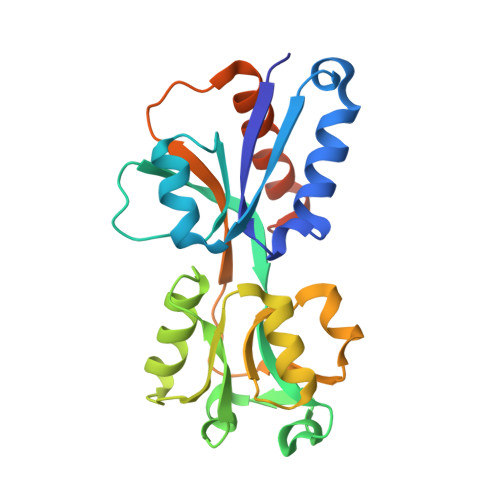
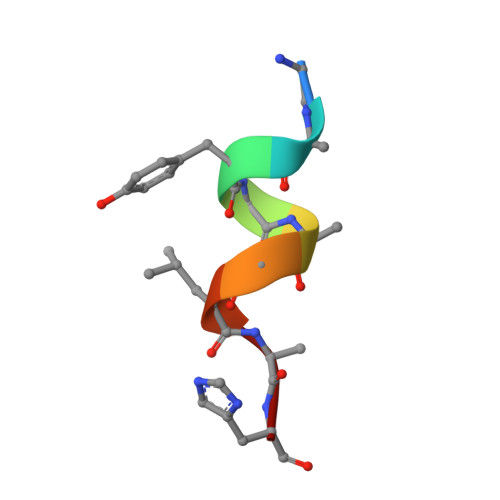
Shikimate Induced Transcriptional Activation of Protocatechuate Biosynthesis Genes by QuiR, a LysR-Type Transcriptional Regulator, in Listeria monocytogenes.
Prezioso, S.M., Xue, K., Leung, N., Gray-Owen, S.D., Christendat, D.(2018) J Mol Biol 430: 1265-1283
- PubMed: 29530613
- DOI: https://doi.org/10.1016/j.jmb.2018.03.003
- Primary Citation of Related Structures:
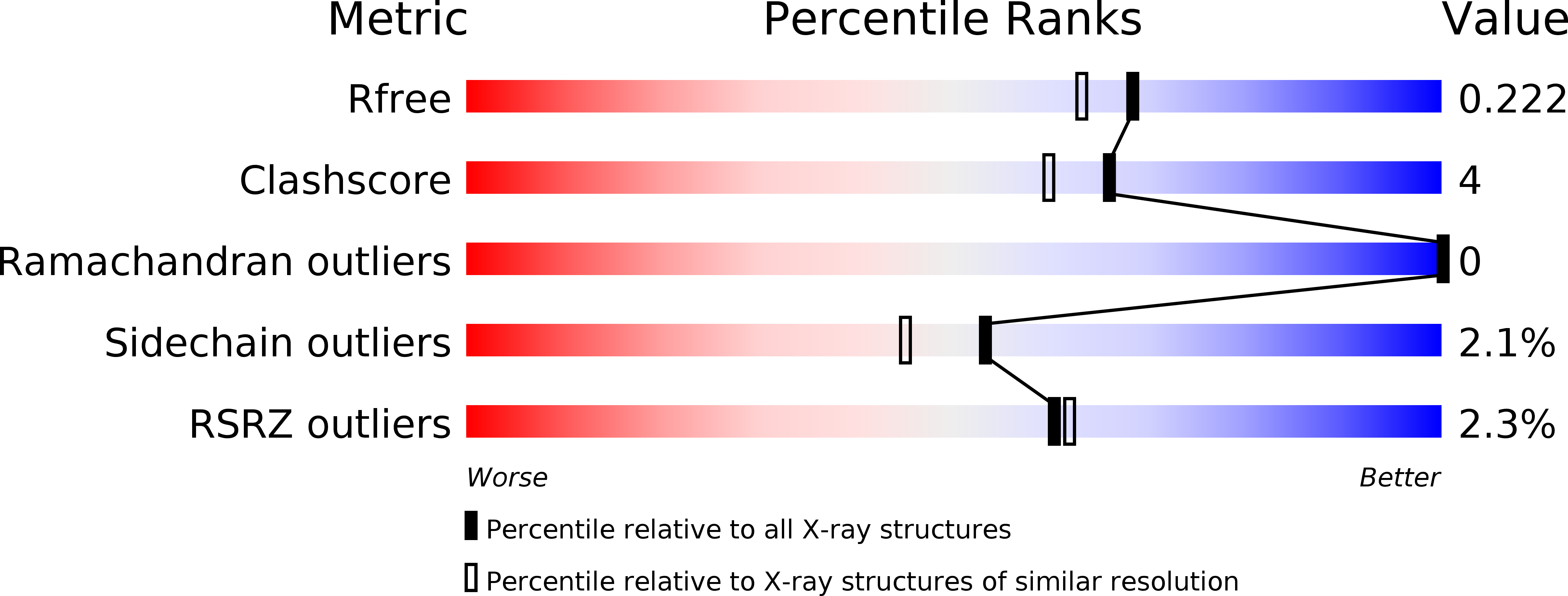
5TED - PubMed Abstract:
Listeria monocytogenes is a common foodborne bacterial pathogen that contaminates plant and animal consumable products. The persistent nature of L. monocytogenes is associated with millions of dollars in food recalls annually. Here, we describe the role of shikimate in directly modulating the expression of genes encoding enzymes for the conversion of quinate and shikimate metabolites to protocatechuate. In L. monocytogenes, these genes are found within two operons, named qui1 and qui2. In addition, a gene named quiR, encoding a LysR-Type Transcriptional Regulator (QuiR), is located immediately upstream of the qui1 operon. Transcriptional lacZ-promoter fusion experiments show that QuiR induces gene expression of both qui1 and qui2 operons in the presence of shikimate. Furthermore, co-crystallization of the QuiR effector binding domain in complex with shikimate provides insights into the mechanism of activation of this regulator. Together these data show that upon shikimate accumulation, QuiR activates the transcription of genes encoding enzymes involved in shikimate and quinate utilization for the production of protocatechuate. Furthermore, the accumulation of protocatechuate leads to the inhibition of Listeria growth. Since protocatechuate is not known to be utilized by Listeria, its role is distinct from those established in other bacteria.
Organizational Affiliation:
Department of Cell and Systems Biology, University of Toronto, 25 Willcocks Street, Toronto, Ontario, Canada M5S 3B2.