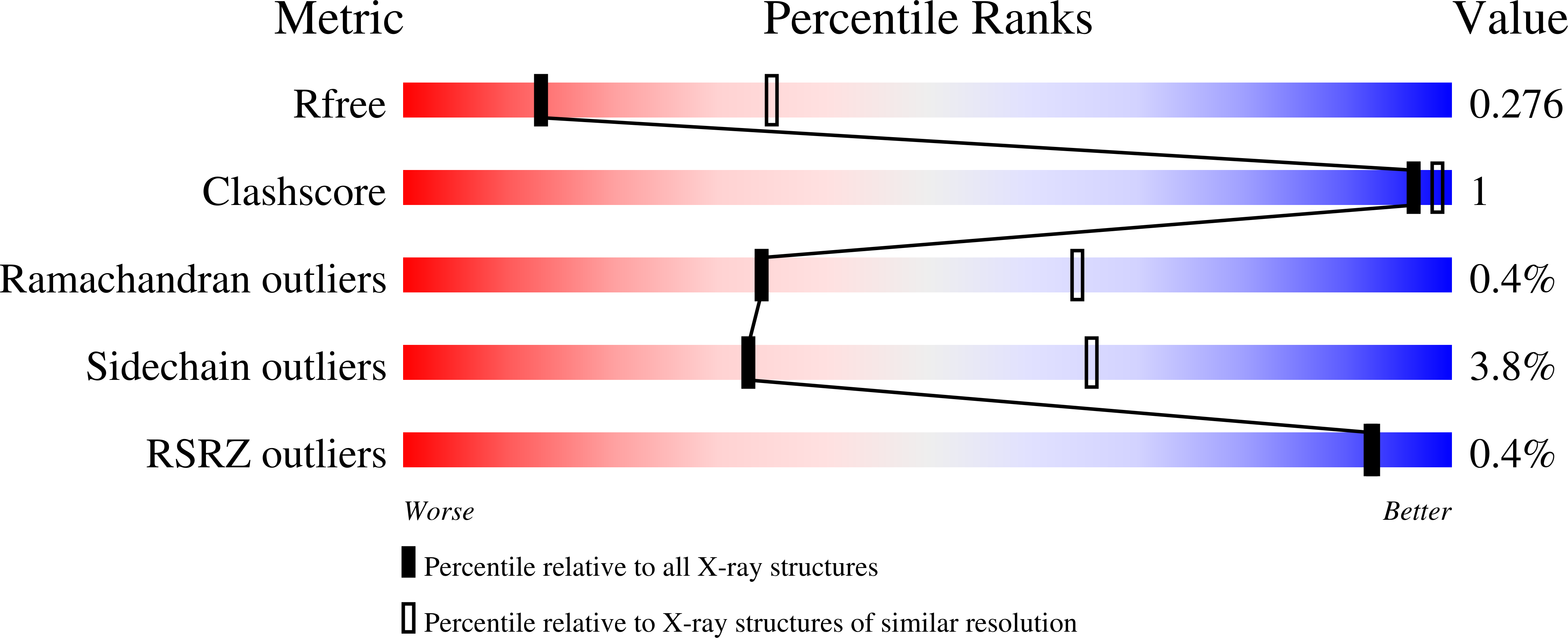
Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
Taylor, A.B., Roberts, K.M., Cao, X., Clark, N.E., Holloway, S.P., Donati, E., Polcaro, C.M., Pica-Mattoccia, L., Tarpley, R.S., McHardy, S.F., Cioli, D., LoVerde, P.T., Fitzpatrick, P.F., Hart, P.J.(2017) J Biol Chem 292: 11154-11164
- PubMed: 28536265
- DOI: https://doi.org/10.1074/jbc.M116.766527
- Primary Citation of Related Structures:
5TIV, 5TIW, 5TIX, 5TIY, 5TIZ - PubMed Abstract:
The antischistosomal prodrug oxamniquine is activated by a sulfotransferase (SULT) in the parasitic flatworm Schistosoma mansoni. Of the three main human schistosome species, only S. mansoni is sensitive to oxamniquine therapy despite the presence of SULT orthologs in Schistosoma hematobium and Schistosoma japonicum The reason for this species-specific drug action has remained a mystery for decades. Here we present the crystal structures of S. hematobium and S. japonicum SULTs, including S. hematobium SULT in complex with oxamniquine. We also examined the activity of the three enzymes in vitro ; surprisingly, all three are active toward oxamniquine, yet we observed differences in catalytic efficiency that implicate kinetics as the determinant for species-specific toxicity. These results provide guidance for designing oxamniquine derivatives to treat infection caused by all species of schistosome to combat emerging resistance to current therapy.
Organizational Affiliation:
From the Departments of Biochemistry and Structural Biology and [email protected].