Crystal Structure of 14-3-3 zeta in Complex with a Serine 124-phosphorylated TBC1D7 peptide
HU, J., DONG, A., MADIGAN, J., WALKER, J.R., Bountra, C., Arrowsmith, C.H., Edwards, A.M., TONG, Y., Structural Genomics Consortium (SGC)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 14-3-3 protein zeta/delta | 246 | Homo sapiens | Mutation(s): 0 Gene Names: YWHAZ | 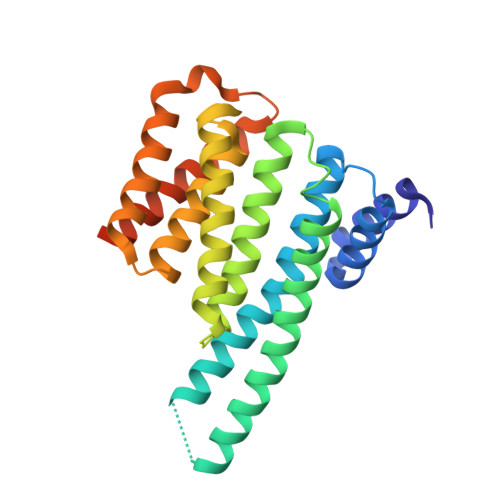 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63104 (Homo sapiens) Explore P63104 Go to UniProtKB: P63104 | |||||
PHAROS: P63104 GTEx: ENSG00000164924 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63104 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TBC1 domain family member 7 peptide | 14 | Homo sapiens | Mutation(s): 2 | 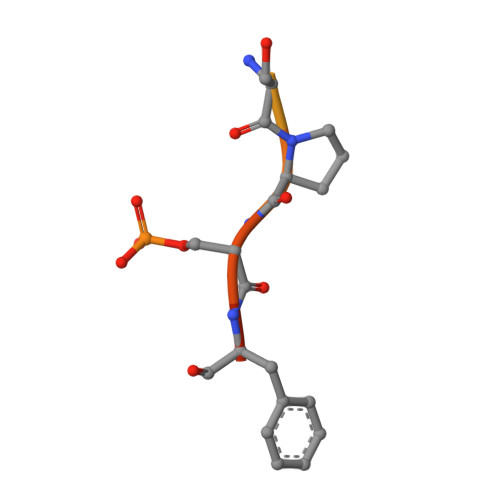 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9P0N9 (Homo sapiens) Explore Q9P0N9 Go to UniProtKB: Q9P0N9 | |||||
PHAROS: Q9P0N9 GTEx: ENSG00000145979 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P0N9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | E [auth A], F [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| UNX Query on UNX | G [auth A] H [auth A] I [auth B] J [auth B] K [auth B] | UNKNOWN ATOM OR ION X |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | C, D | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.709 | α = 90 |
| b = 71.575 | β = 90 |
| c = 129.899 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| BUSTER | refinement |
| PDB_EXTRACT | data extraction |
| PHASER | phasing |
| HKL-3000 | data reduction |