IgG Fc domains that bind C1q but not effector Fc gamma receptors delineate the importance of complement-mediated effector functions.
Lee, C.H., Romain, G., Yan, W., Watanabe, M., Charab, W., Todorova, B., Lee, J., Triplett, K., Donkor, M., Lungu, O.I., Lux, A., Marshall, N., Lindorfer, M.A., Goff, O.R., Balbino, B., Kang, T.H., Tanno, H., Delidakis, G., Alford, C., Taylor, R.P., Nimmerjahn, F., Varadarajan, N., Bruhns, P., Zhang, Y.J., Georgiou, G.(2017) Nat Immunol 18: 889-898
- PubMed: 28604720
- DOI: https://doi.org/10.1038/ni.3770
- Primary Citation of Related Structures:
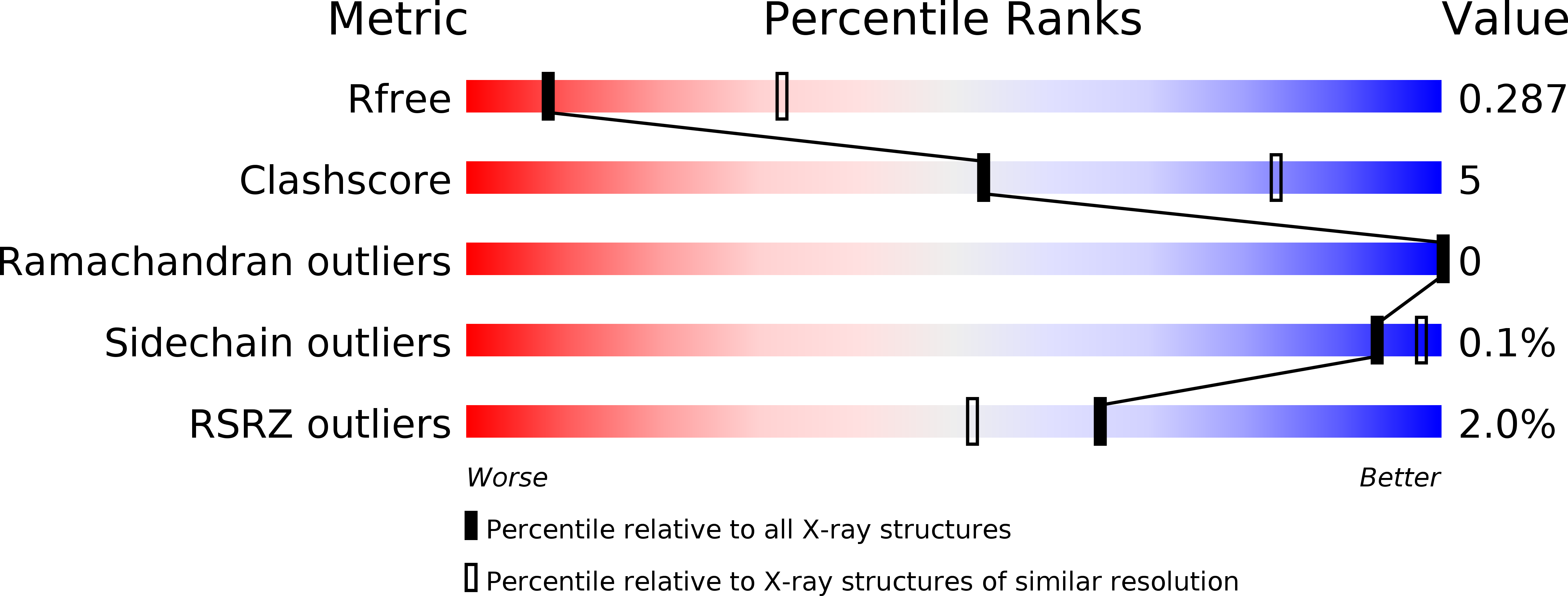
5V43, 5V4E - PubMed Abstract:
Engineered crystallizable fragment (Fc) regions of antibody domains, which assume a unique and unprecedented asymmetric structure within the homodimeric Fc polypeptide, enable completely selective binding to the complement component C1q and activation of complement via the classical pathway without any concomitant engagement of the Fcγ receptor (FcγR). We used the engineered Fc domains to demonstrate in vitro and in mouse models that for therapeutic antibodies, complement-dependent cell-mediated cytotoxicity (CDCC) and complement-dependent cell-mediated phagocytosis (CDCP) by immunological effector molecules mediated the clearance of target cells with kinetics and efficacy comparable to those of the FcγR-dependent effector functions that are much better studied, while they circumvented certain adverse reactions associated with FcγR engagement. Collectively, our data highlight the importance of CDCC and CDCP in monoclonal-antibody function and provide an experimental approach for delineating the effect of complement-dependent effector-cell engagement in various therapeutic settings.
Organizational Affiliation:
Department of Chemical Engineering, University of Texas at Austin, Austin, Texas, USA.