HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Yasutake, Y., Hattori, S.I., Hayashi, H., Matsuda, K., Tamura, N., Kohgo, S., Maeda, K., Mitsuya, H.(2018) Sci Rep 8: 1624-1624
- PubMed: 29374261
- DOI: https://doi.org/10.1038/s41598-018-19602-9
- Primary Citation of Related Structures:
5XN0, 5XN1, 5XN2 - PubMed Abstract:
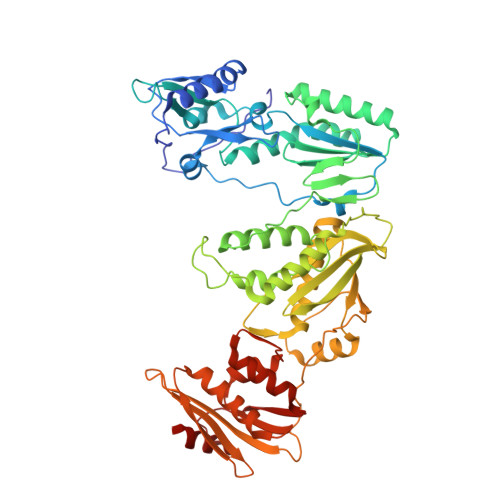
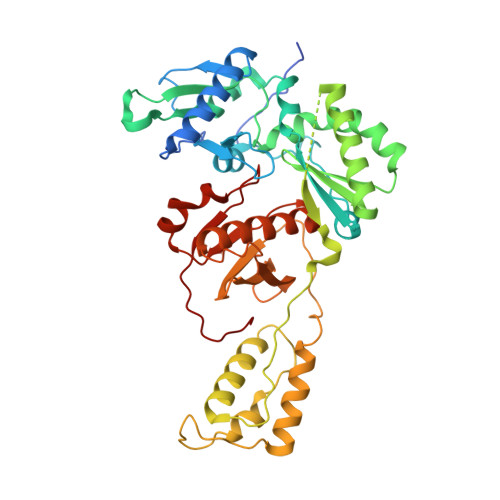
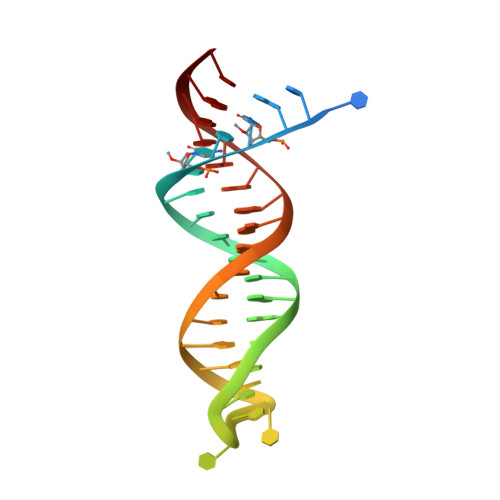
Hepatitis B virus (HBV) reverse transcriptase (RT) is essential for viral replication and is an important drug target. Nonetheless, the notorious insolubility of HBV RT has hindered experimental structural studies and structure-based drug design. Here, we demonstrate that a Q151M substitution alone at the nucleotide-binding site (N-site) of human immunodeficiency virus type-1 (HIV-1) RT renders HIV-1 highly sensitive to entecavir (ETV), a potent nucleoside analogue RT inhibitor (NRTI) against HBV. The results suggest that Met151 forms a transient hydrophobic interaction with the cyclopentyl methylene of ETV, a characteristic hydrophobic moiety of ETV. We thus solved the crystal structures of HIV-1 RT Q151M :DNA complex with bound dGTP or ETV-triphosphate (ETV-TP). The structures revealed that ETV-TP is accommodated at the N-site slightly apart from the ribose ring of the 3'-end nucleotide, compared to the position of bound dGTP and previously reported NRTI/dNTP. In addition, the protruding methylene group of bound ETV-TP directly pushes the side-chain of Met184 backward. Met184 is a key residue that confers ETV resistance upon substitution with smaller Ile/Val. These results provide novel insights into NRTI binding to the N-site and further provide important clues for the development of novel anti-HBV/HIV-1 RT inhibitors to overcome critical drug resistance.
Organizational Affiliation:
Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Sapporo, 062-8517, Japan. [email protected].