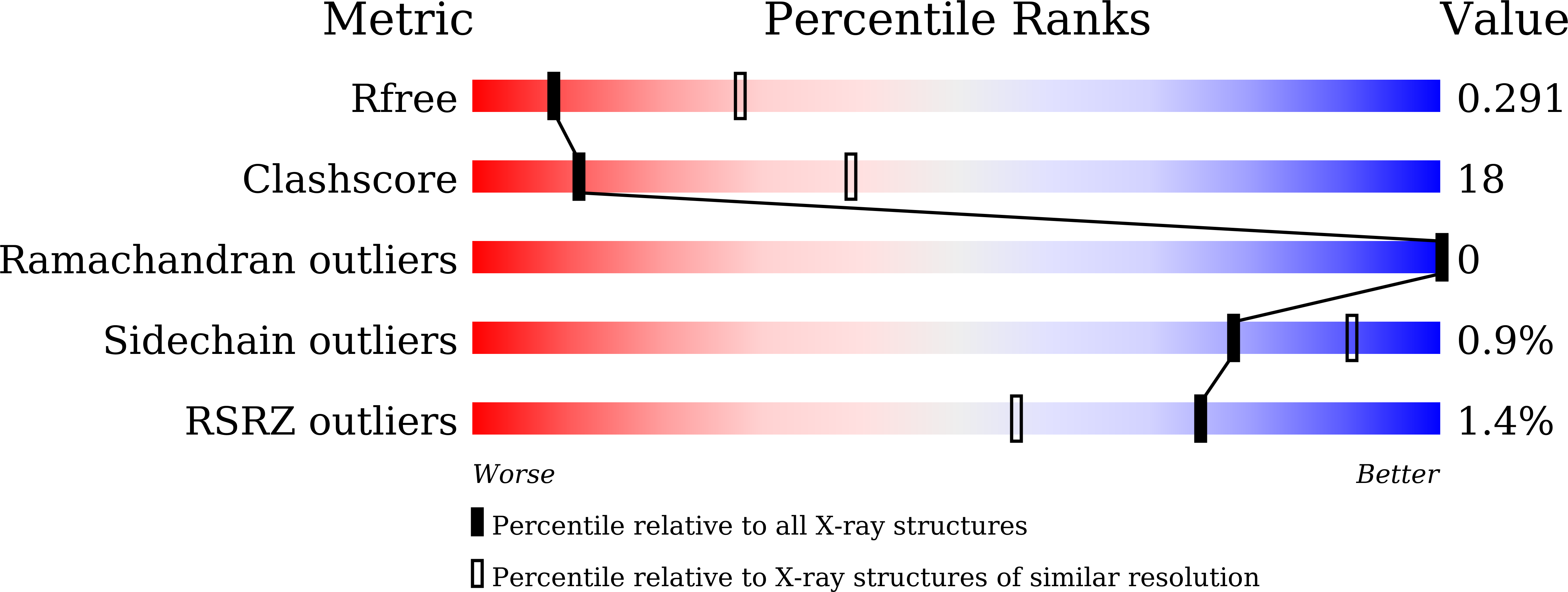
Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Omura, K., Aiba, Y., Onoda, H., Stanfield, J.K., Ariyasu, S., Sugimoto, H., Shiro, Y., Shoji, O., Watanabe, Y.(2018) Chem Commun (Camb) 54: 7892-7895
- PubMed: 29845154
- DOI: https://doi.org/10.1039/c8cc02760a
- Primary Citation of Related Structures:
5ZIS, 5ZLH - PubMed Abstract:
Haem substitution is an effective approach to tweak the function of haemoproteins. Herein, we report a facile haem substitution method for self-sufficient cytochrome P450BM3 (CYP102A1) from Bacillus megaterium utilising the transpeptidase Sortase A from Staphylococcus aureus. We successfully constructed Mn-substituted BM3 and investigated its catalytic activity.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-0802, Japan. [email protected].