Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
Vashisht, S., Kumar, A., Kaur, K.J., Salunke, D.M.(2016) ChemistrySelect 1: 6287-6292
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
A newer entry is available that reflects an alternative modeling of the original data: 5VF2
(2016) ChemistrySelect 1: 6287-6292
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| scFv 2D10 | 251 | Mus musculus | Mutation(s): 0 | 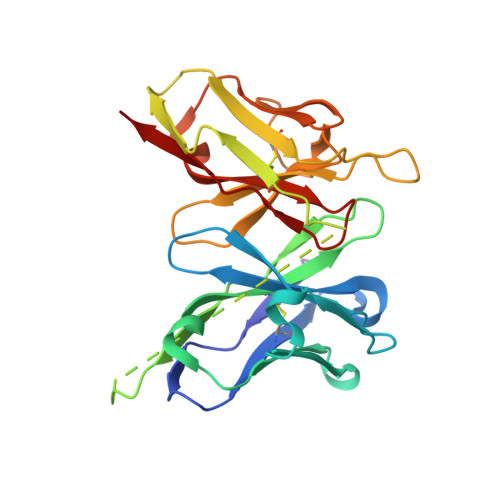 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900118 Query on PRD_900118 | B, C | 6alpha-alpha-mannobiose | Oligosaccharide / Metabolism |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.093 | α = 90 |
| b = 81.093 | β = 90 |
| c = 74.388 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHASER | phasing |