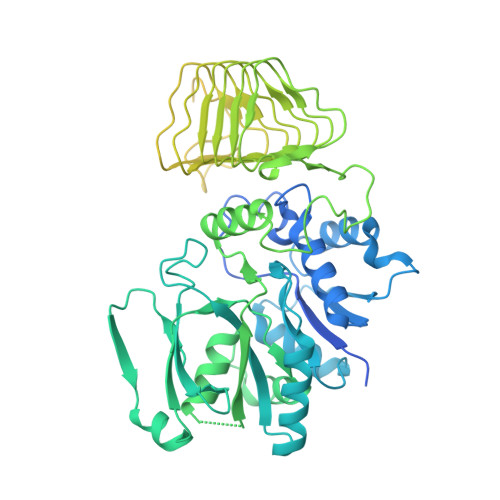
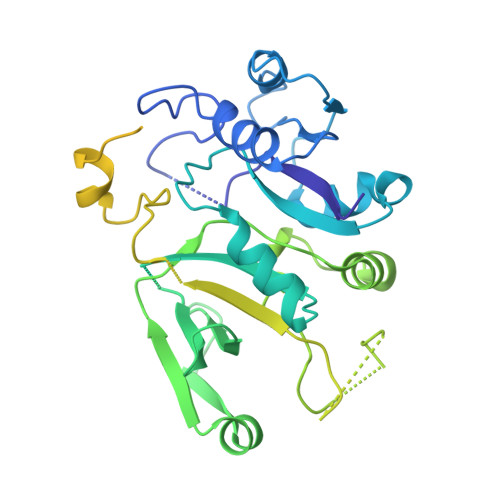
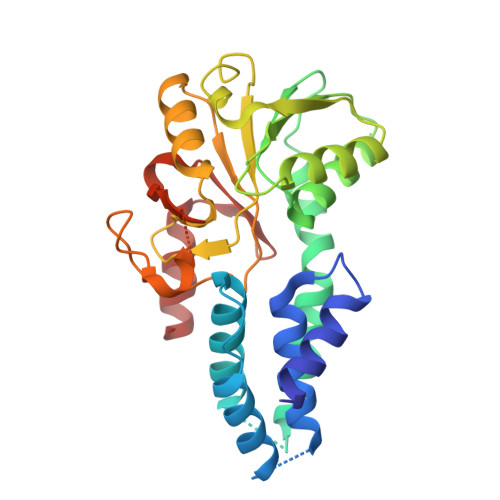
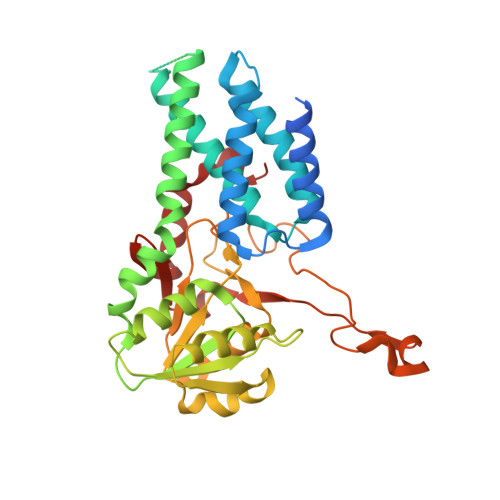
Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule.
Tsai, J.C., Miller-Vedam, L.E., Anand, A.A., Jaishankar, P., Nguyen, H.C., Renslo, A.R., Frost, A., Walter, P.(2018) Science 359
- PubMed: 29599213
- DOI: https://doi.org/10.1126/science.aaq0939
- Primary Citation of Related Structures:
6CAJ - PubMed Abstract:
Regulation by the integrated stress response (ISR) converges on the phosphorylation of translation initiation factor eIF2 in response to a variety of stresses. Phosphorylation converts eIF2 from a substrate to a competitive inhibitor of its dedicated guanine nucleotide exchange factor, eIF2B, thereby inhibiting translation. ISRIB, a drug-like eIF2B activator, reverses the effects of eIF2 phosphorylation, and in rodents it enhances cognition and corrects cognitive deficits after brain injury. To determine its mechanism of action, we solved an atomic-resolution structure of ISRIB bound in a deep cleft within decameric human eIF2B by cryo-electron microscopy. Formation of fully active, decameric eIF2B holoenzyme depended on the assembly of two identical tetrameric subcomplexes, and ISRIB promoted this step by cross-bridging a central symmetry interface. Thus, regulation of eIF2B assembly emerges as a rheostat for eIF2B activity that tunes translation during the ISR and that can be further modulated by ISRIB.
Organizational Affiliation:
Howard Hughes Medical Institute, University of California, San Francisco, CA, USA.