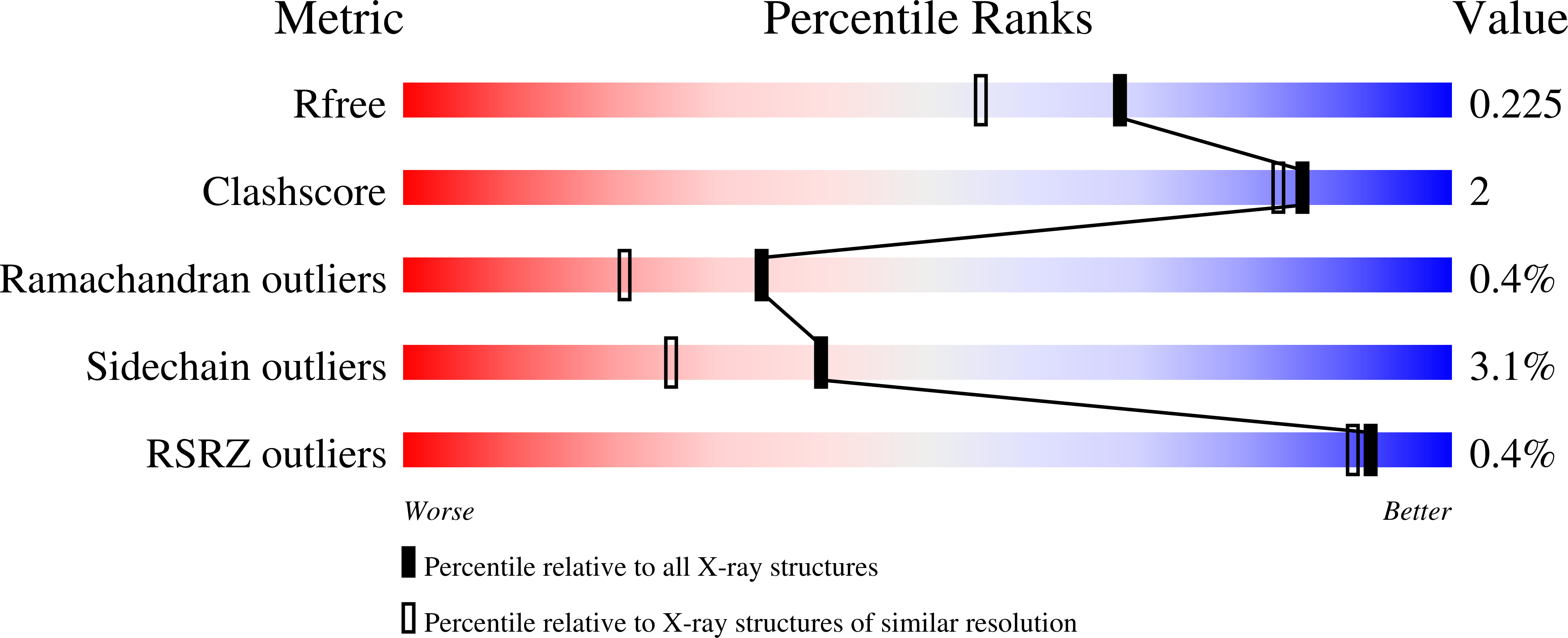
Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Schumann, B., Malaker, S.A., Wisnovsky, S.P., Debets, M.F., Agbay, A.J., Fernandez, D., Wagner, L.J.S., Lin, L., Li, Z., Choi, J., Fox, D.M., Peh, J., Gray, M.A., Pedram, K., Kohler, J.J., Mrksich, M., Bertozzi, C.R.(2020) Mol Cell 78: 824-834.e15
- PubMed: 32325029
- DOI: https://doi.org/10.1016/j.molcel.2020.03.030
- Primary Citation of Related Structures:
6E7I, 6NQT - PubMed Abstract:
Studying posttranslational modifications classically relies on experimental strategies that oversimplify the complex biosynthetic machineries of living cells. Protein glycosylation contributes to essential biological processes, but correlating glycan structure, underlying protein, and disease-relevant biosynthetic regulation is currently elusive. Here, we engineer living cells to tag glycans with editable chemical functionalities while providing information on biosynthesis, physiological context, and glycan fine structure. We introduce a non-natural substrate biosynthetic pathway and use engineered glycosyltransferases to incorporate chemically tagged sugars into the cell surface glycome of the living cell. We apply the strategy to a particularly redundant yet disease-relevant human glycosyltransferase family, the polypeptide N-acetylgalactosaminyl transferases. This approach bestows a gain-of-chemical-functionality modification on cells, where the products of individual glycosyltransferases can be selectively characterized or manipulated to understand glycan contribution to major physiological processes.
Organizational Affiliation:
Department of Chemistry, Stanford University, Stanford, CA 94305, USA; Chemical Glycobiology Laboratory, The Francis Crick Institute, NW1 1AT London, United Kingdom; Department of Chemistry, Imperial College London, W12 0BZ London, United Kingdom. Electronic address: [email protected].