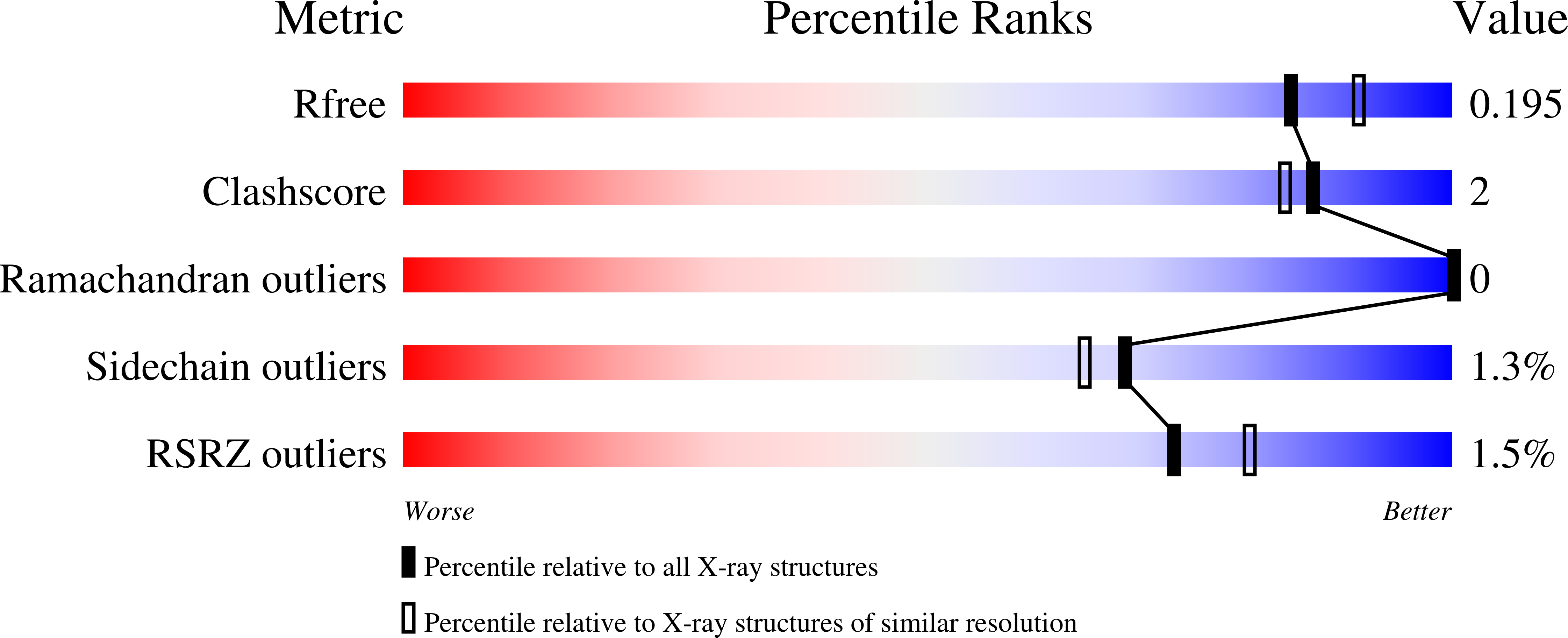
Feasibility and physiological relevance of designing highly potent aminopeptidase-sparing leukotriene A4 hydrolase inhibitors.
Numao, S., Hasler, F., Laguerre, C., Srinivas, H., Wack, N., Jager, P., Schmid, A., Osmont, A., Rothlisberger, P., Houguenade, J., Bergsdorf, C., Dawson, J., Carte, N., Hofmann, A., Markert, C., Hardaker, L., Billich, A., Wolf, R.M., Penno, C.A., Bollbuck, B., Miltz, W., Rohn, T.A.(2017) Sci Rep 7: 13591-13591
- PubMed: 29051536
- DOI: https://doi.org/10.1038/s41598-017-13490-1
- Primary Citation of Related Structures:
6ENB, 6ENC, 6END - PubMed Abstract:
Leukotriene A4 Hydrolase (LTA4H) is a bifunctional zinc metalloenzyme that comprises both epoxide hydrolase and aminopeptidase activity, exerted by two overlapping catalytic sites. The epoxide hydrolase function of the enzyme catalyzes the biosynthesis of the pro-inflammatory lipid mediator leukotriene (LT) B4. Recent literature suggests that the aminopeptidase function of LTA4H is responsible for degradation of the tripeptide Pro-Gly-Pro (PGP) for which neutrophil chemotactic activity has been postulated. It has been speculated that the design of epoxide hydrolase selective LTA4H inhibitors that spare the aminopeptidase pocket may therefore lead to more efficacious anti-inflammatory drugs. In this study, we conducted a high throughput screen (HTS) for LTA4H inhibitors and attempted to rationally design compounds that would spare the PGP degrading function. While we were able to identify compounds with preference for the epoxide hydrolase function, absolute selectivity was not achievable for highly potent compounds. In order to assess the relevance of designing such aminopeptidase-sparing LTA4H inhibitors, we studied the role of PGP in inducing inflammation in different settings in wild type and LTA4H deficient (LTA4H KO) animals but could not confirm its chemotactic potential. Attempting to design highly potent epoxide hydrolase selective LTA4H inhibitors, therefore seems to be neither feasible nor relevant.
Organizational Affiliation:
Chemical Biology & Therapeutics, Novartis Institutes for BioMedical Research, Novartis Pharma AG, Basel, Switzerland.