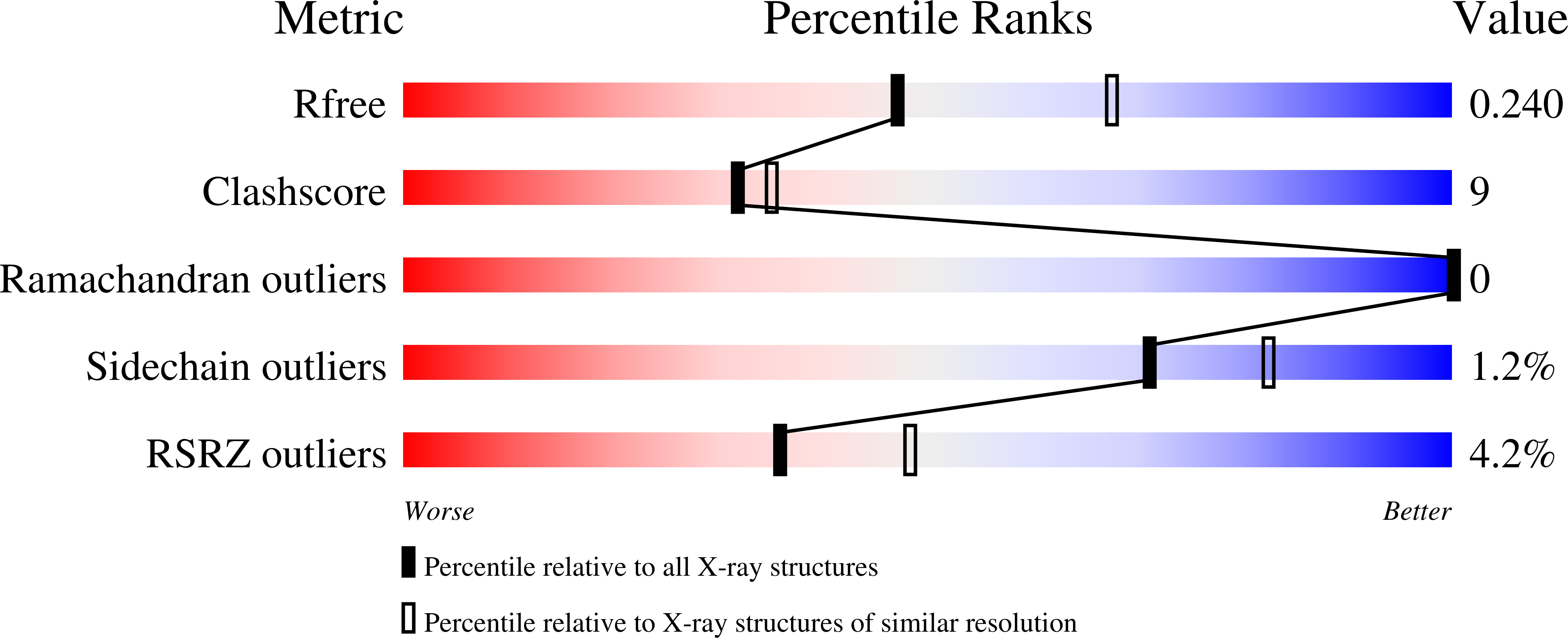
A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Sleiman, D., Garcia, P.S., Lagune, M., Loc'h, J., Haouz, A., Taib, N., Rothlisberger, P., Gribaldo, S., Marliere, P., Kaminski, P.A.(2021) Science 372: 516-520
- PubMed: 33926955
- DOI: https://doi.org/10.1126/science.abe6494
- Primary Citation of Related Structures:
6FLF, 6FM0, 6FM1, 6TNH - PubMed Abstract:
Cells have two purine pathways that synthesize adenine and guanine ribonucleotides from phosphoribose via inosylate. A chemical hybrid between adenine and guanine, 2-aminoadenine (Z), replaces adenine in the DNA of the cyanobacterial virus S-2L. We show that S-2L and Vibrio phage PhiVC8 encode a third purine pathway catalyzed by PurZ, a distant paralog of succinoadenylate synthase (PurA), the enzyme condensing aspartate and inosylate in the adenine pathway. PurZ condenses aspartate with deoxyguanylate into dSMP (N6-succino-2-amino-2'-deoxyadenylate), which undergoes defumarylation and phosphorylation to give dZTP (2-amino-2'-deoxyadenosine-5'-triphosphate), a substrate for the phage DNA polymerase. Crystallography and phylogenetics analyses indicate a close relationship between phage PurZ and archaeal PurA enzymes. Our work elucidates the biocatalytic innovation that remodeled a DNA building block beyond canonical molecular biology.
Organizational Affiliation:
Biology of Gram-Positive Pathogens, Institut Pasteur, CNRS-UMR 2001, Paris, France.