Characterization of Plasmodium falciparum 6-Phosphogluconate Dehydrogenase as an Antimalarial Drug Target.
Haeussler, K., Fritz-Wolf, K., Reichmann, M., Rahlfs, S., Becker, K.(2018) J Mol Biol 430: 4049-4067
- PubMed: 30098336
- DOI: https://doi.org/10.1016/j.jmb.2018.07.030
- Primary Citation of Related Structures:
6FQX, 6FQY, 6FQZ - PubMed Abstract:
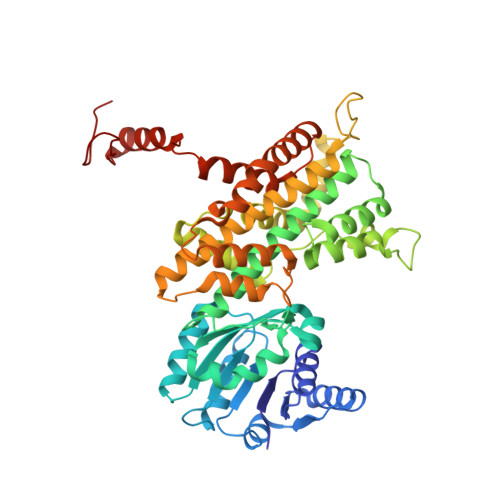
The enzyme 6-phosphogluconate dehydrogenase (6PGD) of the malaria parasite Plasmodium falciparum catalyzes the third step of the pentose phosphate pathway converting 6-phosphogluconate (6PG) to ribulose 5-phosphate. The NADPH produced by 6PGD is crucial for antioxidant defense and redox regulation, and ribose 5-phosphate is essential for DNA and RNA synthesis in the rapidly growing parasite. Thus, 6PGD represents an attractive antimalarial drug target. In this study, we present the X-ray structures of Pf6PGD in native form as well as in complex with 6PG or nicotinamide adenine dinucleotide phosphate (NADP + ) at resolutions of 2.8, 1.9, and 2.9 Å, respectively. The overall structure of the protein is similar to structures of 6PGDs from other species; however, a flexible loop close to the active site rearranges upon binding of 6PG and likely regulates the conformation of the cofactor NADP + . Upon binding of 6PG, the active site loop adopts a closed conformation. In the absence of 6PG, the loop opens and NADP + is bound in a waiting position, indicating that the cofactor and 6PG bind independently from each other. This sequential binding mechanism was supported by kinetic studies on the homodimeric wild-type Pf6PGD. Furthermore, the function of the Plasmodium-specific residue W104L mutant was characterized by site-directed mutagenesis. Notably, the activity of Pf6PGD was found to be post-translationally redox regulated via S-nitrosylation, and screening the Medicines for Malaria Venture Malaria Box identified several compounds with IC 50 s in the low micromolar range. Together with the three-dimensional structure of the protein, this is a promising starting point for further drug discovery approaches.
Organizational Affiliation:
Biochemistry and Molecular Biology, Interdisciplinary Research Center, Justus Liebig University, 35392 Giessen, Germany.