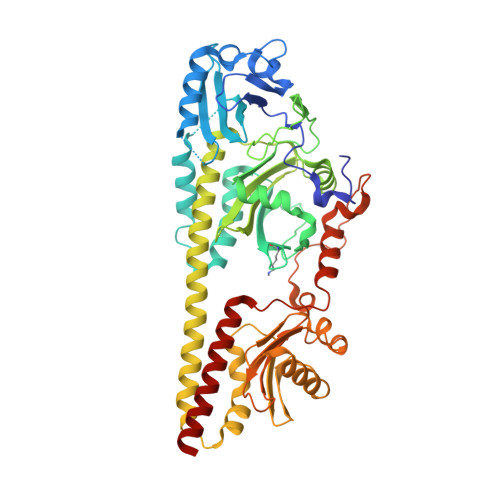
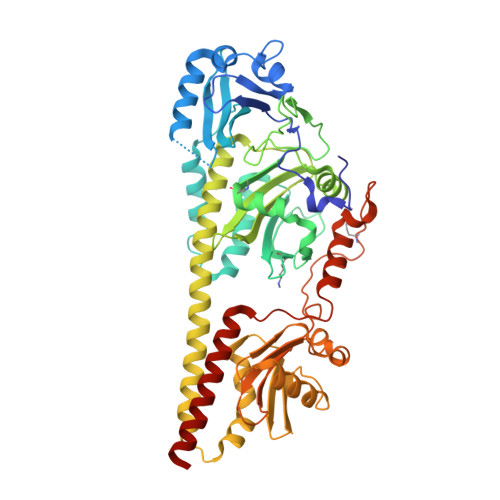
Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Schmidt, A., Sauthof, L., Szczepek, M., Lopez, M.F., Escobar, F.V., Qureshi, B.M., Michael, N., Buhrke, D., Stevens, T., Kwiatkowski, D., von Stetten, D., Mroginski, M.A., Krauss, N., Lamparter, T., Hildebrandt, P., Scheerer, P.(2018) Nat Commun 9: 4912-4912
- PubMed: 30464203
- DOI: https://doi.org/10.1038/s41467-018-07392-7
- Primary Citation of Related Structures:
6G1Y, 6G1Z, 6G20 - PubMed Abstract:
Phytochromes are modular photoreceptors of plants, bacteria and fungi that use light as a source of information to regulate fundamental physiological processes. Interconversion between the active and inactive states is accomplished by a photoinduced reaction sequence which couples the sensor with the output module. However, the underlying molecular mechanism is yet not fully understood due to the lack of structural data of functionally relevant intermediate states. Here we report the crystal structure of a Meta-F intermediate state of an Agp2 variant from Agrobacterium fabrum. This intermediate, the identity of which was verified by resonance Raman spectroscopy, was formed by irradiation of the parent Pfr state and displays significant reorientations of almost all amino acids surrounding the chromophore. Structural comparisons allow identifying structural motifs that might serve as conformational switch for initiating the functional secondary structure change that is linked to the (de-)activation of these photoreceptors.
Organizational Affiliation:
Charité - Universitätsmedizin Berlin, Corporate Member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Institute for Medical Physics and Biophysics, Group Protein X-ray Crystallography and Signal Transduction, Charitéplatz 1, Berlin, D-10117, Germany.