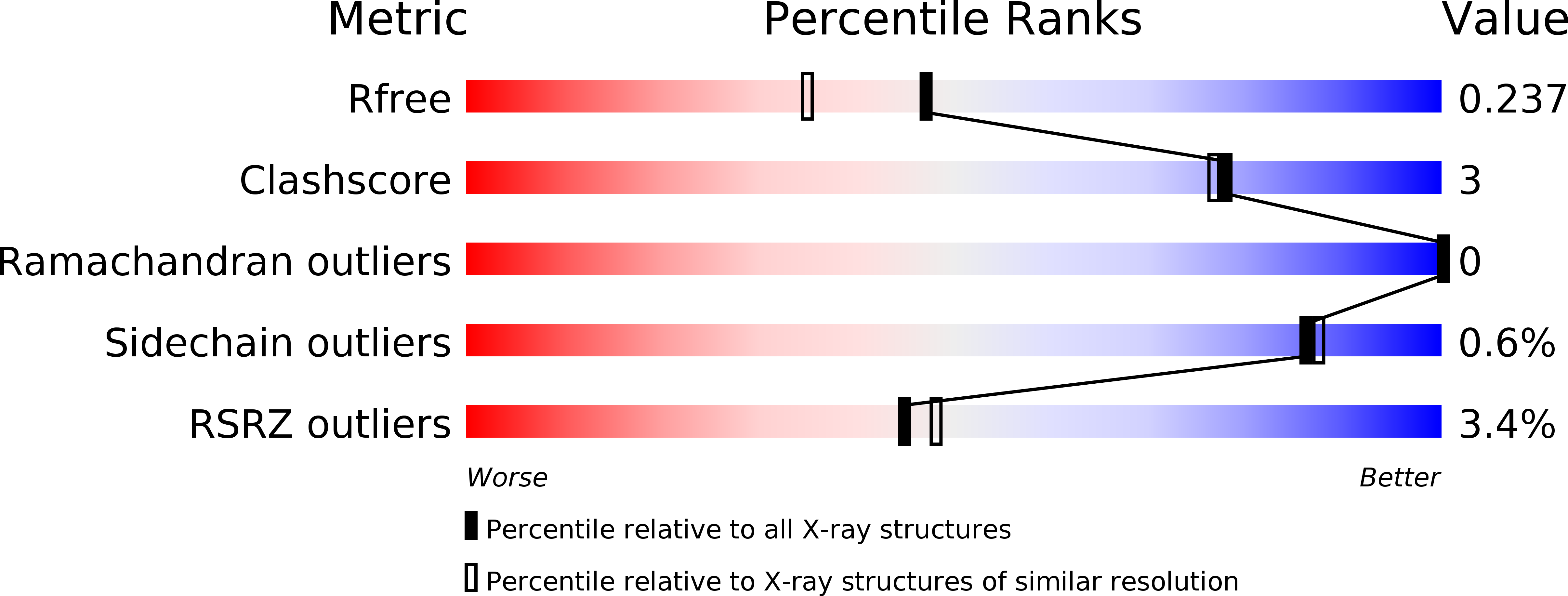
Crystal structures of a beta-trefoil lectin from Entamoeba histolytica in monomeric and a novel disulfide bond-mediated dimeric forms.
Khan, F., Kurre, D., Suguna, K.(2020) Glycobiology 30: 474-488
- PubMed: 31967310
- DOI: https://doi.org/10.1093/glycob/cwaa001
- Primary Citation of Related Structures:
6IFA, 6IFB - PubMed Abstract:
β-Trefoil lectins are galactose/N-acetyl galactosamine specific lectins, which are widely distributed across all kingdoms of life and are known to perform several important functions. However, there is no report available on the characterization of these lectins from protozoans. We have performed structural and biophysical studies on a β-trefoil lectin from Entamoeba histolytica (EntTref), which exists as a mixture of monomers and dimers in solution. Further, we have determined the affinities of EntTref for rhamnose, galactose and different galactose-linked sugars. We obtained the crystal structure of EntTref in a sugar-free form (EntTref_apo) and a rhamnose-bound form (EntTref_rham). A novel Cys residue-mediated dimerization was revealed in the crystal structure of EntTref_apo while the structure of EntTref_rham provided the structural basis for the recognition of rhamnose by a β-trefoil lectin for the first time. To the best of our knowledge, this is the only report of the structural, functional and biophysical characterization of a β-trefoil lectin from a protozoan source and the first report of Cys-mediated dimerization in this class of lectins.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore, CV Raman Rd, 560012, India.