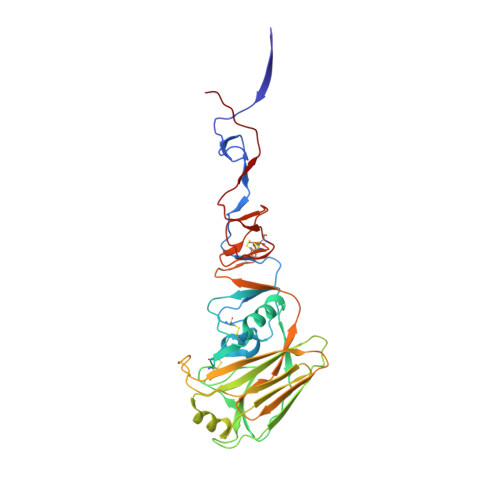
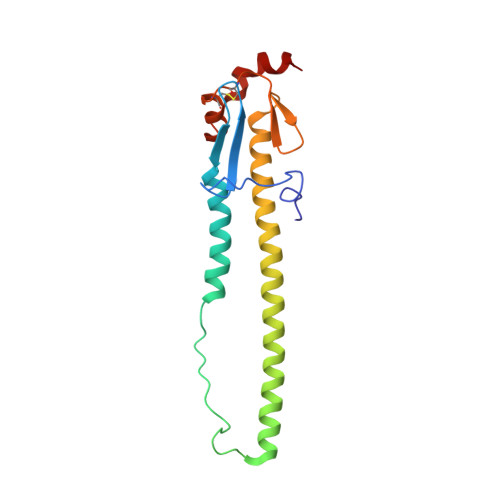
Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Huang, K.A., Rijal, P., Jiang, H., Wang, B., Schimanski, L., Dong, T., Liu, Y.M., Chang, P., Iqbal, M., Wang, M.C., Chen, Z., Song, R., Huang, C.C., Yang, J.H., Qi, J., Lin, T.Y., Li, A., Powell, T.J., Jan, J.T., Ma, C., Gao, G.F., Shi, Y., Townsend, A.R.(2019) Nat Microbiol 4: 306-315
- PubMed: 30478290
- DOI: https://doi.org/10.1038/s41564-018-0303-7
- Primary Citation of Related Structures:
6II4, 6II8, 6II9 - PubMed Abstract:
Little is known about the specificities and neutralization breadth of the H7-reactive antibody repertoire induced by natural H7N9 infection in humans. We have isolated and characterized 73 H7-reactive monoclonal antibodies from peripheral B cells from four donors infected in 2013 and 2014. Of these, 45 antibodies were H7-specific, and 17 of these neutralized the virus, albeit with few somatic mutations in their variable domain sequences. An additional set of 28 antibodies, isolated from younger donors born after 1968, cross-reacted between H7 and H3 haemagglutinins in binding assays, and had accumulated significantly more somatic mutations, but were predominantly non-neutralizing in vitro. Crystal structures of three neutralizing and protective antibodies in complex with the H7 haemagglutinin revealed that they recognize overlapping residues surrounding the receptor-binding site of haemagglutinin. One of the antibodies, L4A-14, bound into the sialic acid binding site and made contacts with haemagglutinin residues that were conserved in the great majority of 2016-2017 H7N9 isolates. However, only 3 of the 17 neutralizing antibodies retained activity for the Yangtze River Delta lineage viruses isolated in 2016-2017 that have undergone antigenic change, which emphasizes the need for updated H7N9 vaccines.
Organizational Affiliation:
Division of Pediatric Infectious Diseases, Department of Pediatrics, Chang Gung Memorial Hospital, Taoyuan, Taiwan. [email protected].