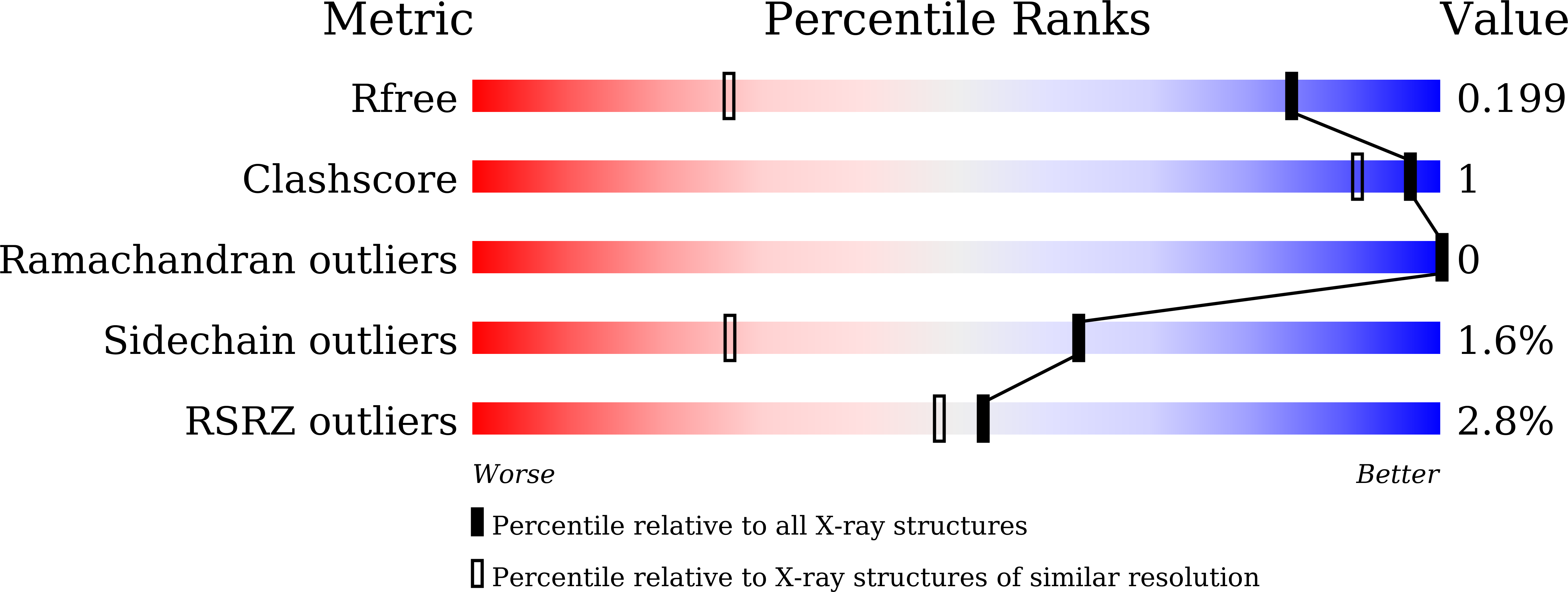
An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Wu, C.Y., Hu, I.C., Yang, Y.C., Ding, W.C., Lai, C.H., Lee, Y.Z., Liu, Y.C., Cheng, H.C., Lyu, P.C.(2020) Commun Biol 3: 441-441
- PubMed: 32796911
- DOI: https://doi.org/10.1038/s42003-020-01177-9
- Primary Citation of Related Structures:
5GI5, 5GI7, 5GI9, 6K80 - PubMed Abstract:
Acetyl coenzyme A (Ac-CoA)-dependent N-acetylation is performed by arylalkylamine N-acetyltransferase (AANAT) and is important in many biofunctions. AANAT catalyzes N-acetylation through an ordered sequential mechanism in which cofactor (Ac-CoA) binds first, with substrate binding afterward. No ternary structure containing AANAT, cofactor, and substrate was determined, meaning the details of substrate binding and product release remain unclear. Here, two ternary complexes of dopamine N-acetyltransferase (Dat) before and after N-acetylation were solved at 1.28 Å and 1.36 Å resolution, respectively. Combined with the structures of Dat in apo form and Ac-CoA bound form, we addressed each stage in the catalytic cycle. Isothermal titration calorimetry (ITC), crystallography, and nuclear magnetic resonance spectroscopy (NMR) were utilized to analyze the product release. Our data revealed that Ac-CoA regulates the conformational properties of Dat to form the catalytic site and substrate binding pocket, while the release of products is facilitated by the binding of new Ac-CoA.
Organizational Affiliation:
Institute of Bioinformatics and Structural Biology, National Tsing Hua University, Hsinchu, 30013, Taiwan.