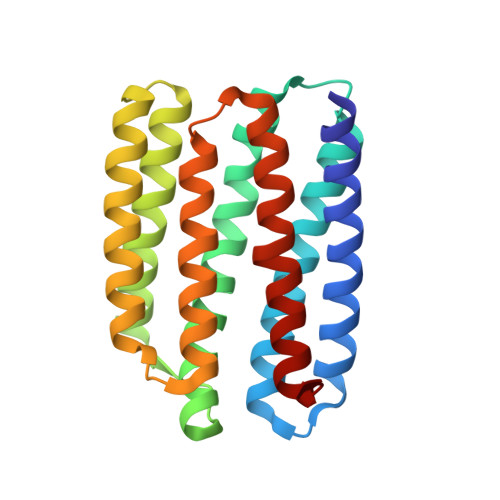
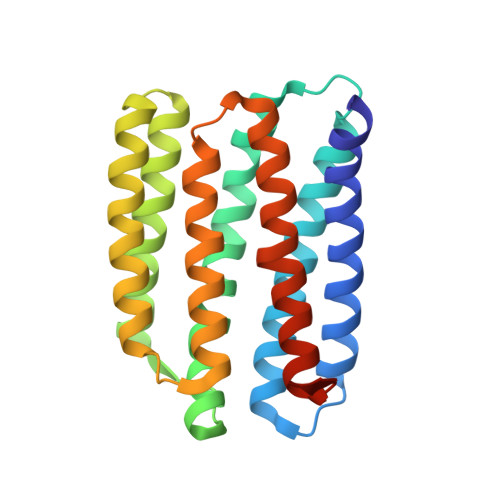
Unique structure and function of viral rhodopsins.
Bratanov, D., Kovalev, K., Machtens, J.P., Astashkin, R., Chizhov, I., Soloviov, D., Volkov, D., Polovinkin, V., Zabelskii, D., Mager, T., Gushchin, I., Rokitskaya, T., Antonenko, Y., Alekseev, A., Shevchenko, V., Yutin, N., Rosselli, R., Baeken, C., Borshchevskiy, V., Bourenkov, G., Popov, A., Balandin, T., Buldt, G., Manstein, D.J., Rodriguez-Valera, F., Fahlke, C., Bamberg, E., Koonin, E., Gordeliy, V.(2019) Nat Commun 10: 4939-4939
- PubMed: 31666521
- DOI: https://doi.org/10.1038/s41467-019-12718-0
- Primary Citation of Related Structures:
6SQG - PubMed Abstract:
Recently, two groups of rhodopsin genes were identified in large double-stranded DNA viruses. The structure and function of viral rhodopsins are unknown. We present functional characterization and high-resolution structure of an Organic Lake Phycodnavirus rhodopsin II (OLPVRII) of group 2. It forms a pentamer, with a symmetrical, bottle-like central channel with the narrow vestibule in the cytoplasmic part covered by a ring of 5 arginines, whereas 5 phenylalanines form a hydrophobic barrier in its exit. The proton donor E42 is placed in the helix B. The structure is unique among the known rhodopsins. Structural and functional data and molecular dynamics suggest that OLPVRII might be a light-gated pentameric ion channel analogous to pentameric ligand-gated ion channels, however, future patch clamp experiments should prove this directly. The data shed light on a fundamentally distinct branch of rhodopsins and may contribute to the understanding of virus-host interactions in ecologically important marine protists.
Organizational Affiliation:
Institute of Complex Systems (ICS), ICS-6: Structural Biochemistry, Forschungszentrum Jülich, Jülich, Germany.