High-resolution structural insights into the heliorhodopsin family.
Kovalev, K., Volkov, D., Astashkin, R., Alekseev, A., Gushchin, I., Haro-Moreno, J.M., Chizhov, I., Siletsky, S., Mamedov, M., Rogachev, A., Balandin, T., Borshchevskiy, V., Popov, A., Bourenkov, G., Bamberg, E., Rodriguez-Valera, F., Buldt, G., Gordeliy, V.(2020) Proc Natl Acad Sci U S A 117: 4131-4141
- PubMed: 32034096
- DOI: https://doi.org/10.1073/pnas.1915888117
- Primary Citation of Related Structures:
6SU3, 6SU4 - PubMed Abstract:
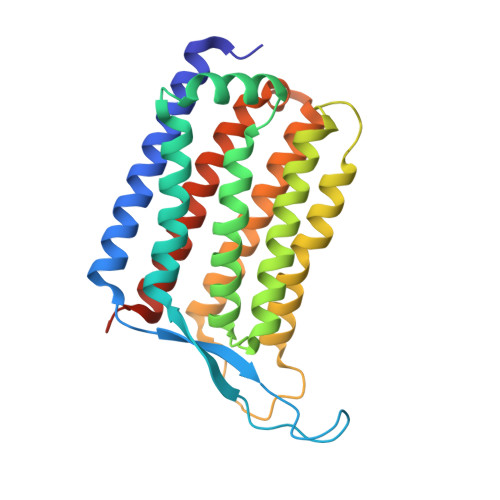
Rhodopsins are the most abundant light-harvesting proteins. A new family of rhodopsins, heliorhodopsins (HeRs), has recently been discovered. Unlike in the known rhodopsins, in HeRs the N termini face the cytoplasm. The function of HeRs remains unknown. We present the structures of the bacterial HeR-48C12 in two states at the resolution of 1.5 Å, which highlight its remarkable difference from all known rhodopsins. The interior of HeR's extracellular part is completely hydrophobic, while the cytoplasmic part comprises a cavity (Schiff base cavity [SBC]) surrounded by charged amino acids and containing a cluster of water molecules, presumably being a primary proton acceptor from the Schiff base. At acidic pH, a planar triangular molecule (acetate) is present in the SBC. Structure-based bioinformatic analysis identified 10 subfamilies of HeRs, suggesting their diverse biological functions. The structures and available data suggest an enzymatic activity of HeR-48C12 subfamily and their possible involvement in fundamental redox biological processes.
Organizational Affiliation:
Institut de Biologie Structurale J.-P. Ebel, Université Grenoble Alpes-Commission for Atomic Energy (CEA)-CNRS, 38000 Grenoble, France.