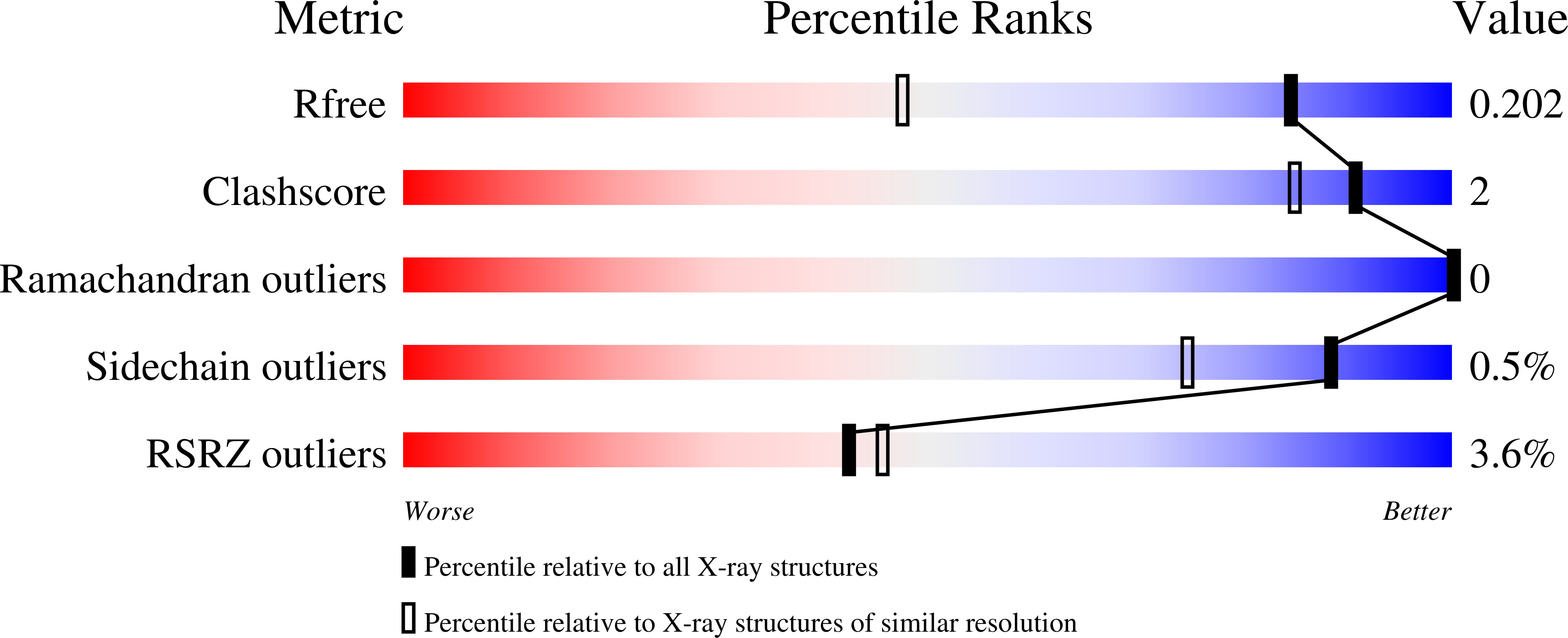
Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Yu, W., Liu, J., Yu, Y., Zhang, V., Clausen, D., Kelly, J., Wolkenberg, S., Beshore, D., Duffy, J.L., Chung, C.C., Myers, R.W., Klein, D.J., Fells, J., Holloway, K., Wu, J., Wu, G., Howell, B.J., Barnard, R.J.O., Kozlowski, J.(2020) Bioorg Med Chem Lett 30: 127197-127197
- PubMed: 32331932
- DOI: https://doi.org/10.1016/j.bmcl.2020.127197
- Primary Citation of Related Structures:
6WBW, 6WBZ - PubMed Abstract:
A novel series of ethyl ketone based HDACs 1, 2, and 3 selective inhibitors have been identified with good enzymatic and cellular activity and high selectivity over HDACs 6 and 8. These inhibitors contain a spirobicyclic group in the amide region. Compound 13 stands out as a lead due to its good potency, high selectivity, and reasonable rat and dog PK. Compounds 33 and 34 show good potency and rat PK profiles as well.
Organizational Affiliation:
Merck & Co., Inc., 2000 Galloping Hill Road, Kenilworth, NJ 07033, USA. Electronic address: [email protected].