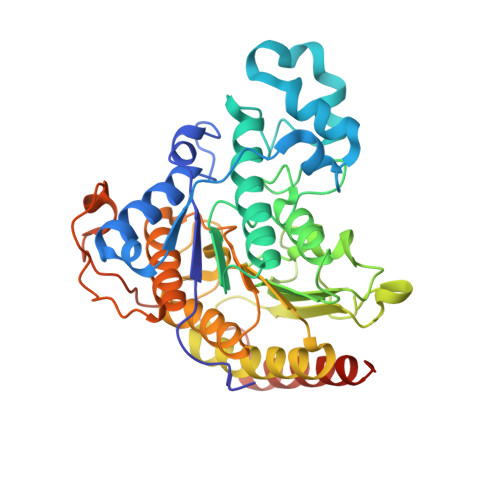
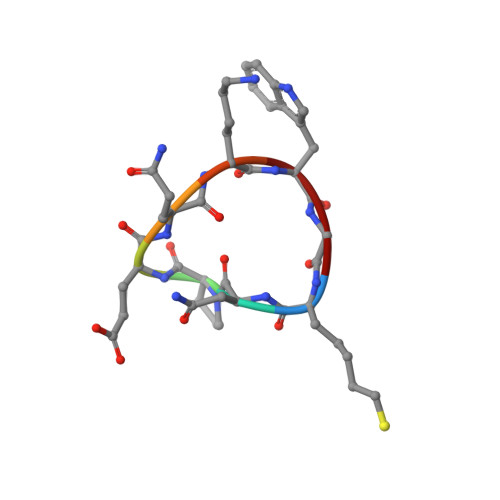
Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Hosseinzadeh, P., Watson, P.R., Craven, T.W., Li, X., Rettie, S., Pardo-Avila, F., Bera, A.K., Mulligan, V.K., Lu, P., Ford, A.S., Weitzner, B.D., Stewart, L.J., Moyer, A.P., Di Piazza, M., Whalen, J.G., Greisen, P.J., Christianson, D.W., Baker, D.(2021) Nat Commun 12: 3384-3384
- PubMed: 34099674
- DOI: https://doi.org/10.1038/s41467-021-23609-8
- Primary Citation of Related Structures:
6WHN, 6WHO, 6WHQ, 6WHZ, 6WI3, 6WSJ - PubMed Abstract:
Despite recent success in computational design of structured cyclic peptides, de novo design of cyclic peptides that bind to any protein functional site remains difficult. To address this challenge, we develop a computational "anchor extension" methodology for targeting protein interfaces by extending a peptide chain around a non-canonical amino acid residue anchor. To test our approach using a well characterized model system, we design cyclic peptides that inhibit histone deacetylases 2 and 6 (HDAC2 and HDAC6) with enhanced potency compared to the original anchor (IC 50 values of 9.1 and 4.4 nM for the best binders compared to 5.4 and 0.6 µM for the anchor, respectively). The HDAC6 inhibitor is among the most potent reported so far. These results highlight the potential for de novo design of high-affinity protein-peptide interfaces, as well as the challenges that remain.
Organizational Affiliation:
University of Washington, Department of Biochemistry, Institute for Protein Design, Seattle, WA, USA.