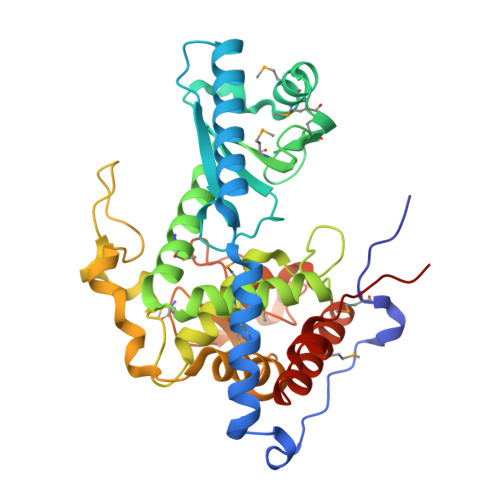
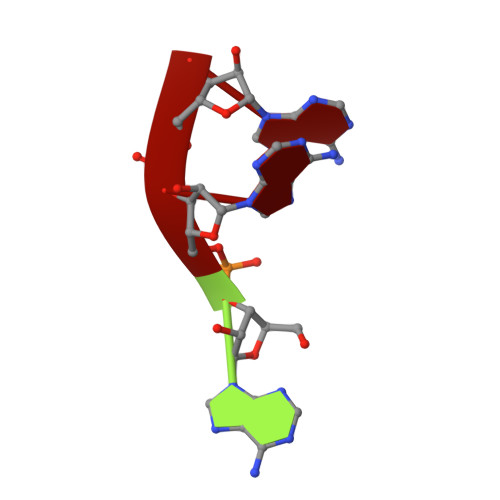
Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Malik, D., Kobylecki, K., Krawczyk, P., Poznanski, J., Jakielaszek, A., Napiorkowska, A., Dziembowski, A., Tomecki, R., Nowotny, M.(2020) Nucleic Acids Res 48: 9387-9405
- PubMed: 32785623
- DOI: https://doi.org/10.1093/nar/gkaa647
- Primary Citation of Related Structures:
6YWN, 6YWO, 6YWP - PubMed Abstract:
Template-independent terminal ribonucleotide transferases (TENTs) catalyze the addition of nucleotide monophosphates to the 3'-end of RNA molecules regulating their fate. TENTs include poly(U) polymerases (PUPs) with a subgroup of 3' CUCU-tagging enzymes, such as CutA in Aspergillus nidulans. CutA preferentially incorporates cytosines, processively polymerizes only adenosines and does not incorporate or extend guanosines. The basis of this peculiar specificity remains to be established. Here, we describe crystal structures of the catalytic core of CutA in complex with an incoming non-hydrolyzable CTP analog and an RNA with three adenosines, along with biochemical characterization of the enzyme. The binding of GTP or a primer with terminal guanosine is predicted to induce clashes between 2-NH2 of the guanine and protein, which would explain why CutA is unable to use these ligands as substrates. Processive adenosine polymerization likely results from the preferential binding of a primer ending with at least two adenosines. Intriguingly, we found that the affinities of CutA for the CTP and UTP are very similar and the structures did not reveal any apparent elements for specific NTP binding. Thus, the properties of CutA likely result from an interplay between several factors, which may include a conformational dynamic process of NTP recognition.
Organizational Affiliation:
Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, Trojdena 4, Warsaw 02-109, Poland.