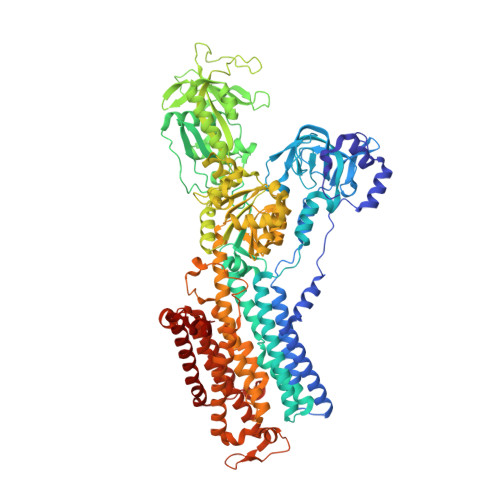
A single K + -binding site in the crystal structure of the gastric proton pump.
Yamamoto, K., Dubey, V., Irie, K., Nakanishi, H., Khandelia, H., Fujiyoshi, Y., Abe, K.(2019) Elife 8
- PubMed: 31436534
- DOI: https://doi.org/10.7554/eLife.47701
- Primary Citation of Related Structures:
6JXH, 6JXI, 6JXJ, 6JXK - PubMed Abstract:
The gastric proton pump (H + ,K + -ATPase), a P-type ATPase responsible for gastric acidification, mediates electro-neutral exchange of H + and K + coupled with ATP hydrolysis, but with an as yet undetermined transport stoichiometry. Here we show crystal structures at a resolution of 2.5 Å of the pump in the E2-P transition state, in which the counter-transporting cation is occluded. We found a single K + bound to the cation-binding site of the H + ,K + -ATPase, indicating an exchange of 1H + /1K + per hydrolysis of one ATP molecule. This fulfills the energy requirement for the generation of a six pH unit gradient across the membrane. The structural basis of K + recognition is resolved and supported by molecular dynamics simulations, establishing how the H + ,K + -ATPase overcomes the energetic challenge to generate an H + gradient of more than a million-fold-one of the highest cation gradients known in mammalian tissue-across the membrane.
Organizational Affiliation:
Cellular and Structural Physiology Institute, Nagoya University, Nagoya, Japan.