Revisiting high-resolution crystal structure of Phormidium rubidum phycocyanin.
Sonani, R.R., Roszak, A.W., Liu, H., Gross, M.L., Blankenship, R.E., Madamwar, D., Cogdell, R.J.(2020) Photosynth Res 144: 349-360
- PubMed: 32303893
- DOI: https://doi.org/10.1007/s11120-020-00746-7
- Primary Citation of Related Structures:
6XWK - PubMed Abstract:
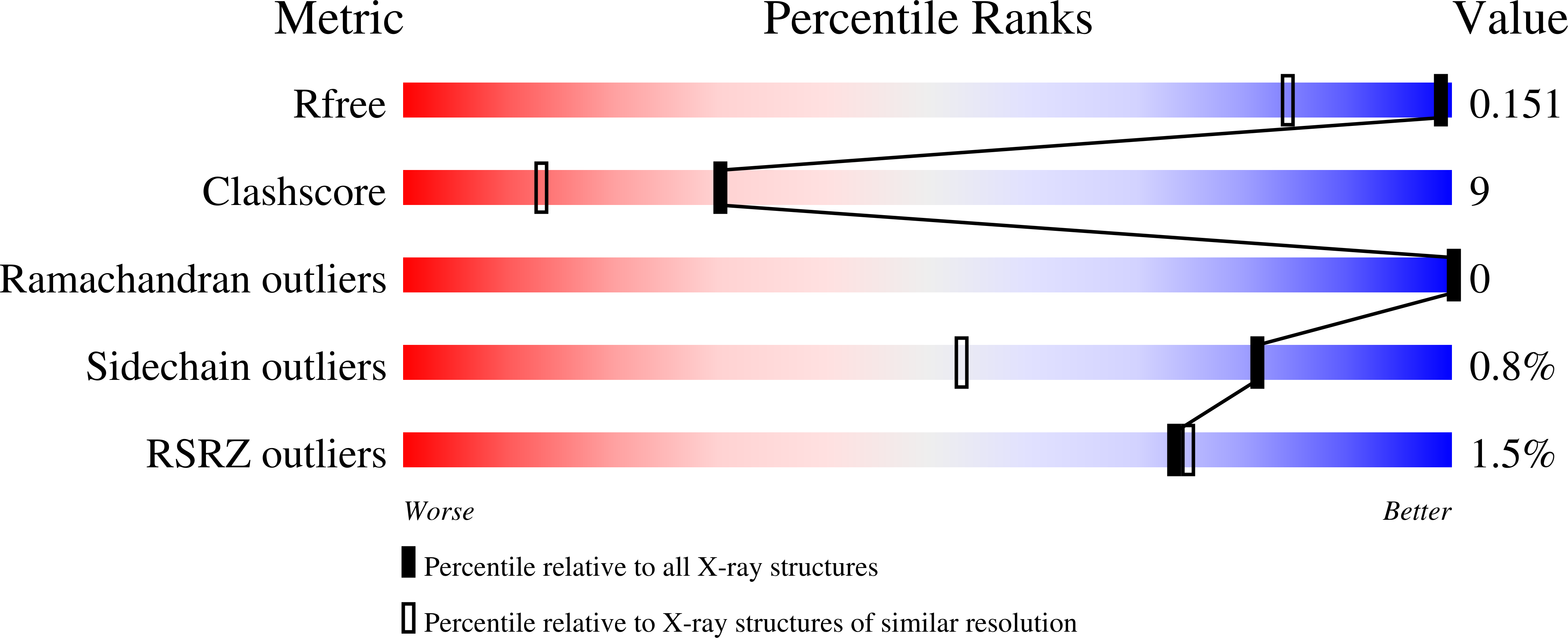
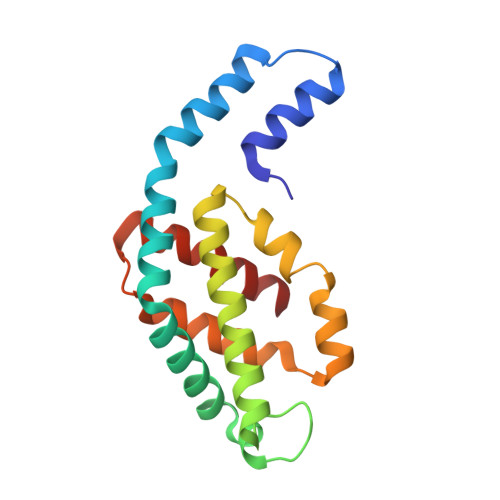
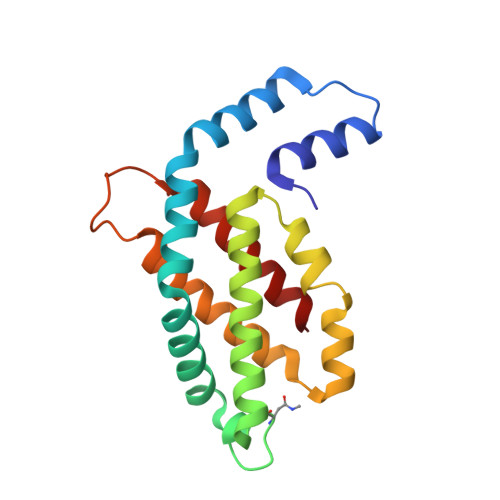
The crystal structure of phycocyanin (pr-PC) isolated from Phormidium rubidum A09DM (P. rubidum) is described at a resolution of 1.17 Å. Electron density maps derived from crystallographic data showed many clear differences in amino acid sequences when compared with the previously obtained gene-derived sequences. The differences were found in 57 positions (30 in α-subunit and 27 in β-subunit of pr-PC), in which all residues except one (β145Arg) are not interacting with the three phycocyanobilin chromophores. Highly purified pr-PC was then sequenced by mass spectrometry (MS) using LC-MS/MS. The MS data were analyzed using two independent proteomic search engines. As a result of this analysis, complete agreement between the polypeptide sequences and the electron density maps was obtained. We attribute the difference to multiple genes in the bacterium encoding the phycocyanin apoproteins and that the gene sequencing sequenced the wrong ones. We are not implying that protein sequencing by mass spectrometry is more accurate than that of gene sequencing. The final 1.17 Å structure of pr-PC allows the chromophore interactions with the protein to be described with high accuracy.
Organizational Affiliation:
Post-Graduate Department of Biosciences, Sardar Patel University, Bakrol, Anand, Gujarat, 388 315, India.