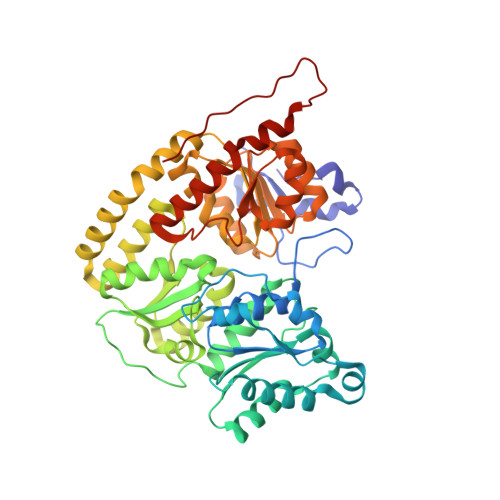
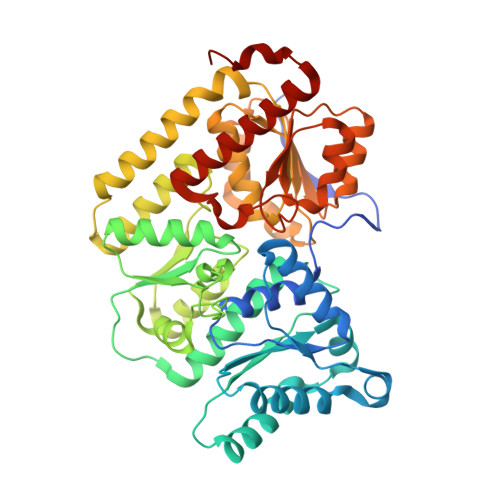
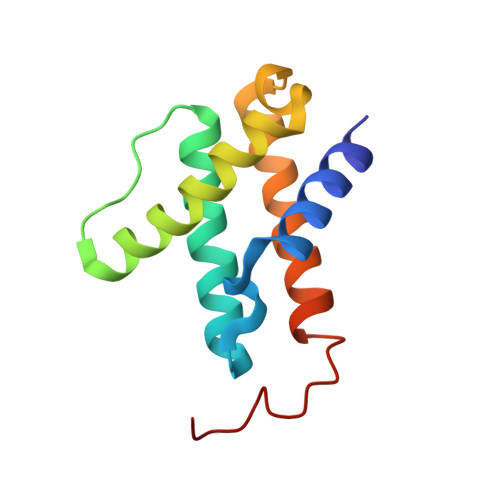
CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Rohde, M., Grunau, K., Einsle, O.(2020) Angew Chem Int Ed Engl 59: 23626-23630
- PubMed: 32915491
- DOI: https://doi.org/10.1002/anie.202010790
- Primary Citation of Related Structures:
7ADR, 7ADY - PubMed Abstract:
Nitrogenases reduce N 2 , the most abundant element in Earth's atmosphere that is otherwise resistant to chemical conversions due to its stable triple bond. Vanadium nitrogenase stands out in that it additionally processes carbon monoxide, a known inhibitor of the reduction of all substrates other than H + . The reduction of CO leads to the formation of hydrocarbon products, holding the potential for biotechnological applications in analogy to the industrial Fischer-Tropsch process. Here we report the most highly resolved structure of vanadium nitrogenase to date at 1.0 Å resolution, with CO bound to the active site cofactor after catalytic turnover. CO bridges iron ions Fe2 and Fe6, replacing sulfide S2B, in a binding mode that is in line with previous reports on the CO complex of molybdenum nitrogenase. We discuss the structural consequences of continued turnover when CO is removed, which involve the replacement of CO possibly by OH - , the movement of Q176 D and K361 D , the return of sulfide and the emergence of two additional water molecules that are absent in the CO-bound state.
Organizational Affiliation:
Institut für Biochemie, Fakultät für Chemie und Pharmazie, Albert-Ludwigs-Universität Freiburg, Albertstrasse 21, 79104, Freiburg im Breisgau, Germany.