Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements.
Panchal, V., Kumar, S., Hossain, S.N., Vasudevan, D.(2021) Int J Biol Macromol 167: 1168-1175
- PubMed: 33197475
- DOI: https://doi.org/10.1016/j.ijbiomac.2020.11.071
- Primary Citation of Related Structures:
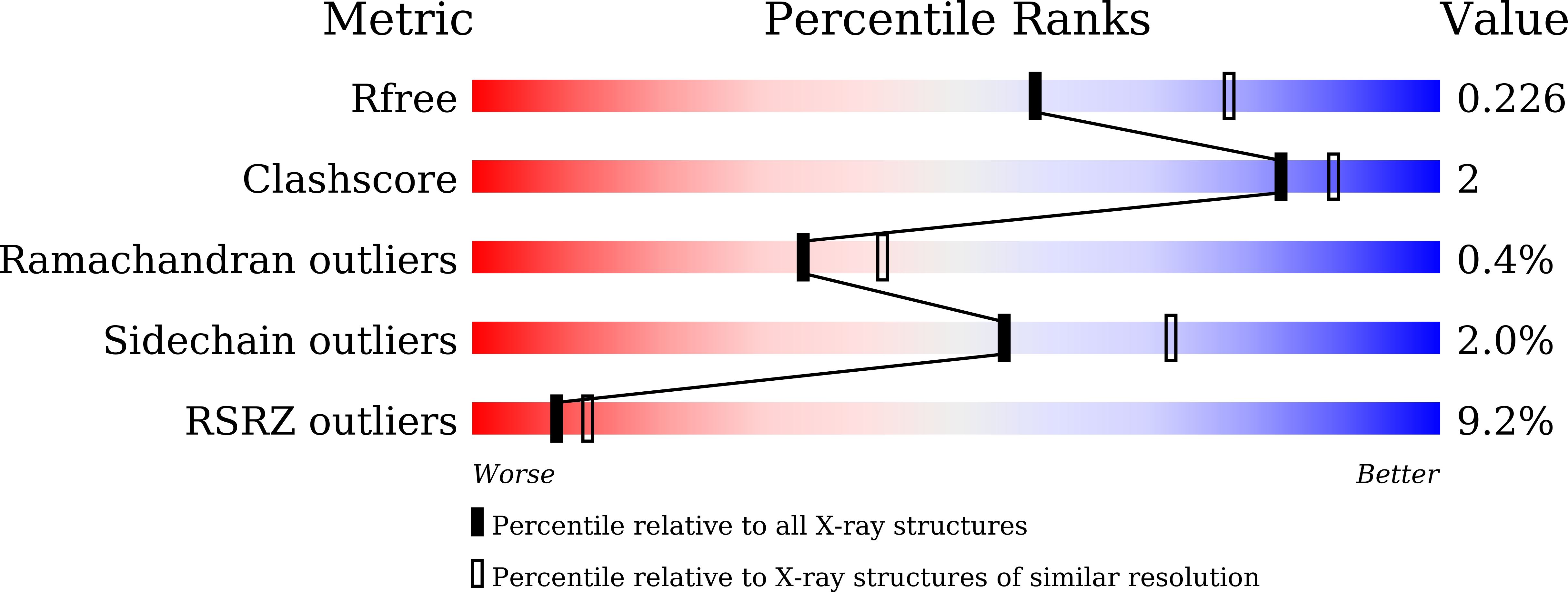
7CC8, 7CCA - PubMed Abstract:
White spot syndrome virus (WSSV), the causative agent of white spot disease (WSD) severely affecting crustacean life forms, is highly contagious and forms the principal cause of massive economic losses in the shrimp aquaculture industry. Previous studies have demonstrated thymidylate synthase as a successful anti-cancer therapeutic drug target, leading to various anti-cancer drugs. The differential utilization of nucleotide precursors between white spot syndrome virus and shrimp encouraged us to analyze WSSV-thymidylate synthase (wTS). Here, we report the crystal structures of wTS in its apo-form and as a ternary complex with deoxyuridine monophosphate (dUMP) and methotrexate at a resolution of 2.35 Å and 2.6 Å, respectively. wTS possesses a fold characteristic to known thymidylate synthase (TS) structures. Like other TS structures, the apo-form of wTS displays an open conformation, whereas the wTS ternary complex attains a closed conformation. While the C-terminal loop maintains a typical distance from methotrexate, the Sγ atom of the catalytic Cys is positioned farther from the C6 atom of dUMP. Altogether, we report the first TS structure from a crustacean virus and highlight its distinction from shrimp and other TS structures.
Organizational Affiliation:
Institute of Life Sciences, Bhubaneswar 751023, Odisha, India.