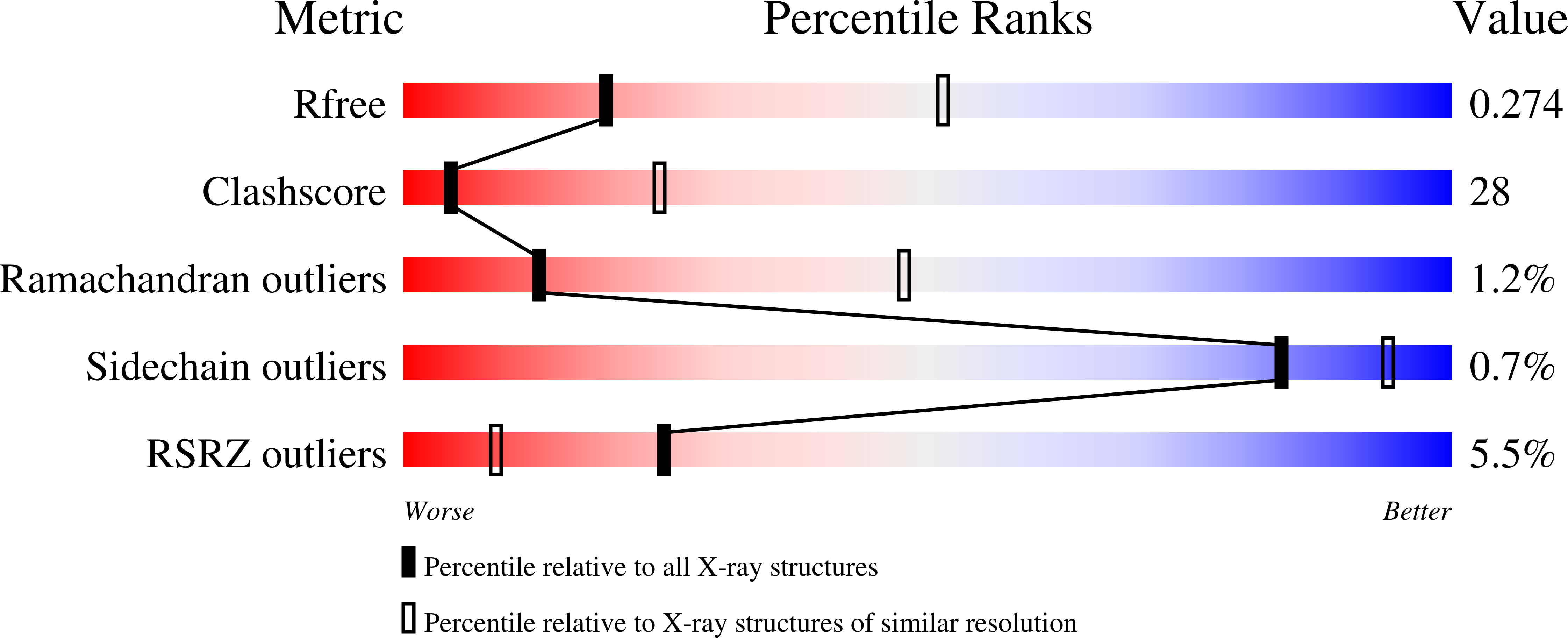
A cell permeable bimane-constrained PCNA-interacting peptide.
Horsfall, A.J., Vandborg, B.A., Kikhtyak, Z., Scanlon, D.B., Tilley, W.D., Hickey, T.E., Bruning, J.B., Abell, A.D.(2021) RSC Chem Biol 2: 1499-1508
- PubMed: 34704055
- DOI: https://doi.org/10.1039/d1cb00113b
- Primary Citation of Related Structures:
7M5L, 7M5M, 7M5N - PubMed Abstract:
The human sliding clamp protein known as proliferating cell nuclear antigen (PCNA) orchestrates DNA-replication and -repair and as such is an ideal therapeutic target for proliferative diseases, including cancer. Peptides derived from the human p21 protein bind PCNA with high affinity via a 3 10 -helical binding conformation and are known to shut down DNA-replication. Here, we present studies on short analogues of p21 peptides (143-151) conformationally constrained with a covalent linker between i , i + 4 separated cysteine residues at positions 145 and 149 to access peptidomimetics that target PCNA. The resulting macrocycles bind PCNA with K D values ranging from 570 nM to 3.86 μM, with the bimane-constrained peptide 7 proving the most potent. Subsequent X-ray crystallography and computational modelling studies of the macrocyclic peptides bound to PCNA indicated only the high-affinity peptide 7 adopted the classical 3 10 -helical binding conformation. This suggests the 3 10 -helical conformation is critical to high affinity PCNA binding, however NMR secondary shift analysis of peptide 7 revealed this secondary structure was not well-defined in solution. Peptide 7 is cell permeable and localised to the cell cytosol of breast cancer cells (MDA-MB-468), revealed by confocal microscopy showing blue fluorescence of the bimane linker. The inherent fluorescence of the bimane moiety present in peptide 7 allowed it to be directly imaged in the cell uptake assay, without attachment of an auxiliary fluorescent tag. This highlights a significant benefit of using a bimane constraint to access conformationally constrained macrocyclic peptides. This study identifies a small peptidomimetic that binds PCNA with higher affinity than previous reported p21 macrocycles, and is cell permeable, providing a significant advance toward development of a PCNA inhibitor for therapeutic applications.
Organizational Affiliation:
Institute of Photonics and Advanced Sensing (IPAS), The University of Adelaide Adelaide South Australia 5005 Australia [email protected] [email protected].