IFN alpha primes cancer cells for Fusicoccin-induced cell death via 14-3-3 PPI stabilization.
Andlovic, B., Heilmann, G., Ninck, S., Andrei, S.A., Centorrino, F., Higuchi, Y., Kato, N., Brunsveld, L., Arkin, M., Menninger, S., Choidas, A., Wolf, A., Klebl, B., Kaschani, F., Kaiser, M., Eickhoff, J., Ottmann, C.(2023) Cell Chem Biol
- PubMed: 37130519
- DOI: https://doi.org/10.1016/j.chembiol.2023.04.005
- Primary Citation of Related Structures:
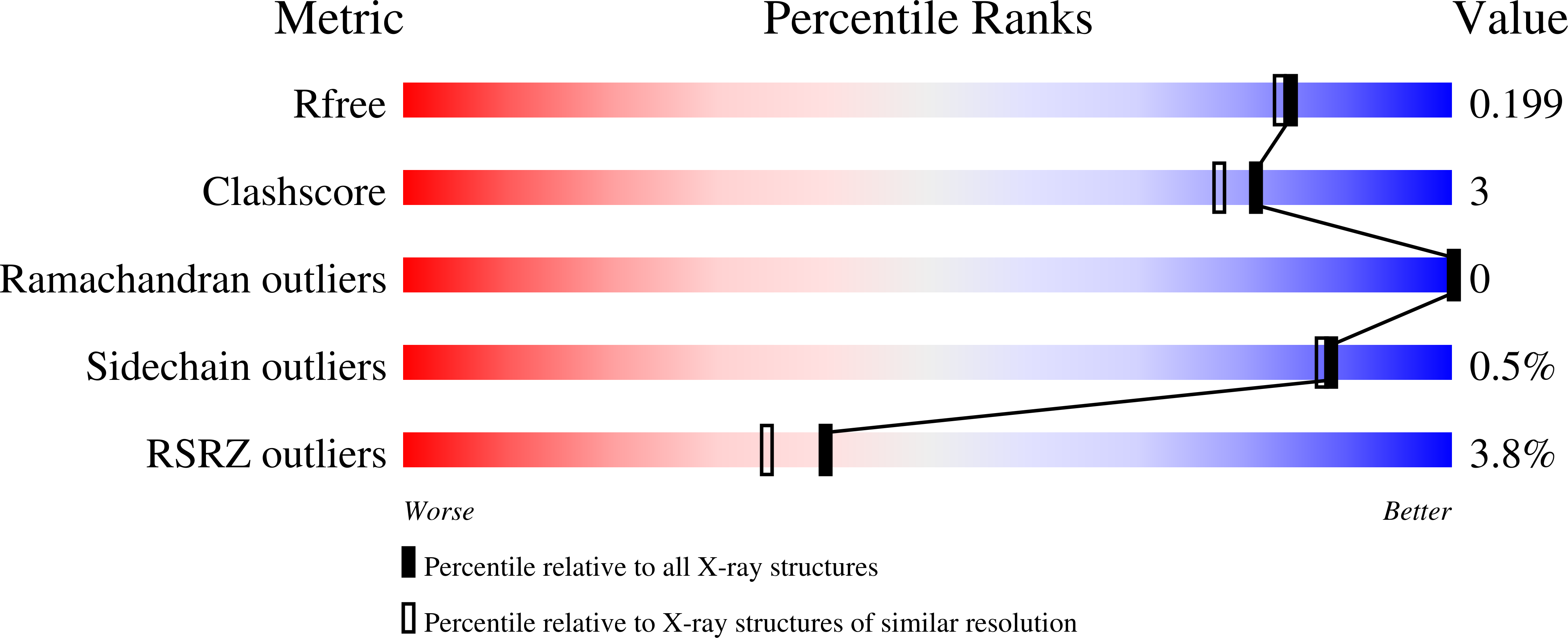
7OB5, 7OB8, 7OBC, 7OBD, 7OBG, 7OBH, 7OBK, 7OBL, 7OBS, 7OBT, 7OBX, 7OBY - PubMed Abstract:
The natural product family of the fusicoccanes (FCs) has been shown to display anti-cancer activity, especially when combined with established therapeutic agents. FCs stabilize 14-3-3 protein-protein interactions (PPIs). Here, we tested combinations of a small library of FCs with interferon α (IFNα) on different cancer cell lines and report a proteomics approach to identify the specific 14-3-3 PPIs that are induced by IFNα and stabilized by FCs in OVCAR-3 cells. Among the identified 14-3-3 target proteins are THEMIS2, receptor interacting protein kinase 2 (RIPK2), EIF2AK2, and several members of the LDB1 complex. Biophysical and structural biology studies confirm these 14-3-3 PPIs as physical targets of FC stabilization, and transcriptome as well as pathway analyses suggest possible explanations for the observed synergistic effect of IFNα/FC treatment on cancer cells. This study elucidates the polypharmacological effects of FCs in cancer cells and identifies potential targets from the vast interactome of 14-3-3s for therapeutic intervention in oncology.
Organizational Affiliation:
Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, Den Dolech 2, 5612 AZ Eindhoven, the Netherlands; Lead Discovery Center GmbH, 44227 Dortmund, Germany.