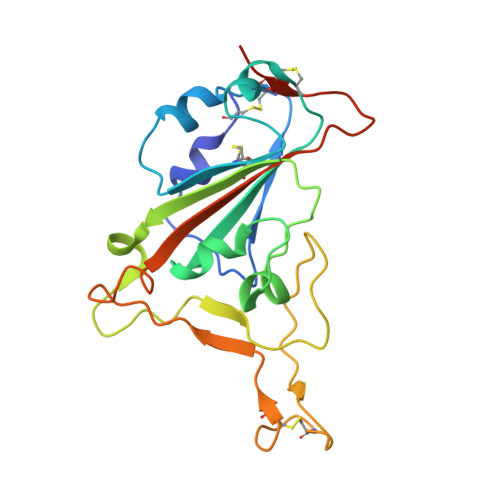
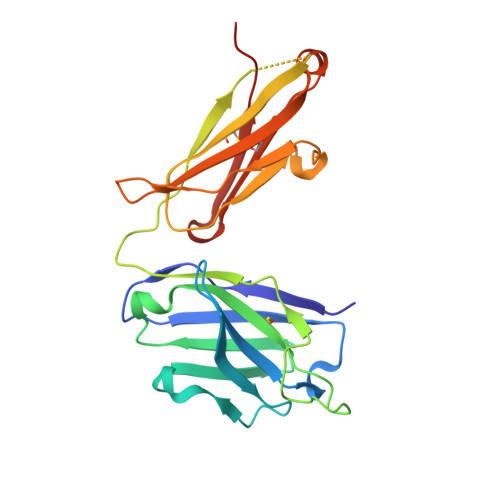
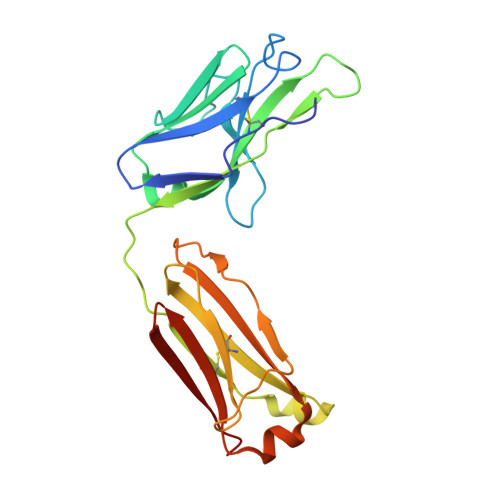
A broad and potent neutralization epitope in SARS-related coronaviruses.
Yuan, M., Zhu, X., He, W.T., Zhou, P., Kaku, C.I., Capozzola, T., Zhu, C.Y., Yu, X., Liu, H., Yu, W., Hua, Y., Tien, H., Peng, L., Song, G., Cottrell, C.A., Schief, W.R., Nemazee, D., Walker, L.M., Andrabi, R., Burton, D.R., Wilson, I.A.(2022) Proc Natl Acad Sci U S A 119: e2205784119-e2205784119
- PubMed: 35767670
- DOI: https://doi.org/10.1073/pnas.2205784119
- Primary Citation of Related Structures:
7U2D, 7U2E - PubMed Abstract:
Many neutralizing antibodies (nAbs) elicited to ancestral severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) through natural infection and vaccination have reduced effectiveness to SARS-CoV-2 variants. Here, we show that therapeutic antibody ADG20 is able to neutralize SARS-CoV-2 variants of concern (VOCs) including Omicron (B.1.1.529) as well as other SARS-related coronaviruses. We delineate the structural basis of this relatively escape-resistant epitope that extends from one end of the receptor binding site (RBS) into the highly conserved CR3022 site. ADG20 can then benefit from high potency through direct competition with ACE2 in the more variable RBS and interaction with the more highly conserved CR3022 site. Importantly, antibodies that are able to target this site generally neutralize a broad range of VOCs, albeit with reduced potency against Omicron. Thus, this conserved and vulnerable site can be exploited for the design of universal vaccines and therapeutic antibodies.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037.