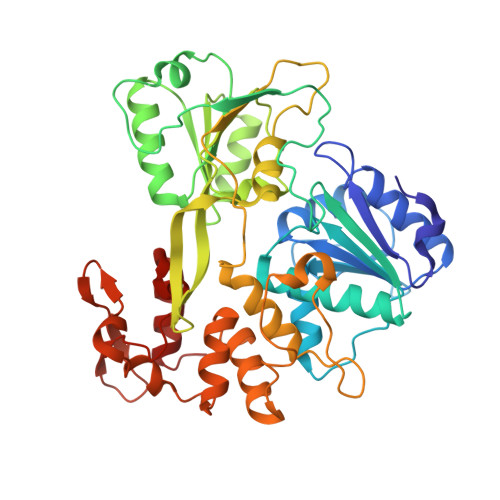
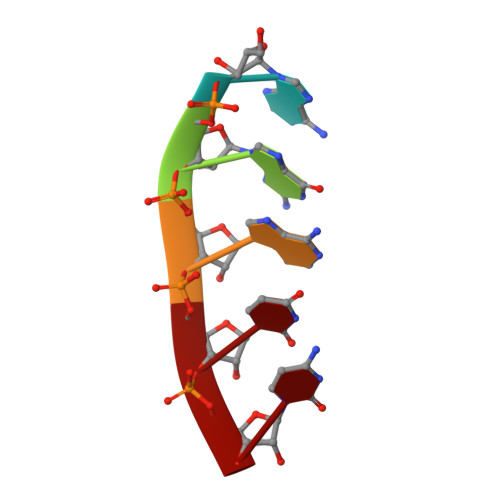
Structural Basis of Zika Virus Helicase in RNA Unwinding and ATP Hydrolysis.
Lin, M., Cui, W., Tian, H., Zhang, Y., Chen, C., Yang, X., Chi, H., Mu, Z., Chen, C., Wang, Z., Ji, X., Yang, H., Lin, Z.(2022) ACS Infect Dis 8: 150-158
- PubMed: 34904824
- DOI: https://doi.org/10.1021/acsinfecdis.1c00455
- Primary Citation of Related Structures:
7V2Z - PubMed Abstract:
The flavivirus nonstructural protein 3 helicase (NS3hel) is a multifunctional domain protein that is associated with DNA/RNA helicase, nucleoside triphosphatase (NTPase), and RNA 5'-triphosphatase (RTPase) activities. As an NTPase-dependent superfamily 2 (SF2) member, NS3hel employs an NTP-driven motor force to unwind double-stranded RNA while translocating along single-stranded RNA and is extensively involved in the viral replication process. Although the structures of SF2 helicases are widely investigated as promising drug targets, the mechanism of energy transduction between NTP hydrolysis and the RNA binding sites in ZIKV NS3hel remains elusive. Here, we report the crystal structure of ZIKV NS3hel in complex with its natural substrates ATP-Mn 2+ and ssRNA. Distinct from other members of the Flavivirus genus, ssRNA binding to ZIKV NS3hel induces relocation of the active water molecules and ATP-associated metal ions in the NTP hydrolysis active site, which promotes the hydrolysis of ATP and the production of AMP. Our findings highlight the importance of the allosteric role of ssRNA on the modulation of ATP hydrolysis and energy utilization.
Organizational Affiliation:
School of Life Sciences, Tianjin University, Tianjin 300072, China.