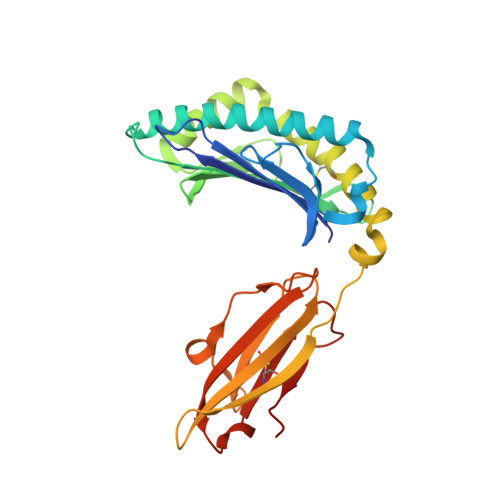
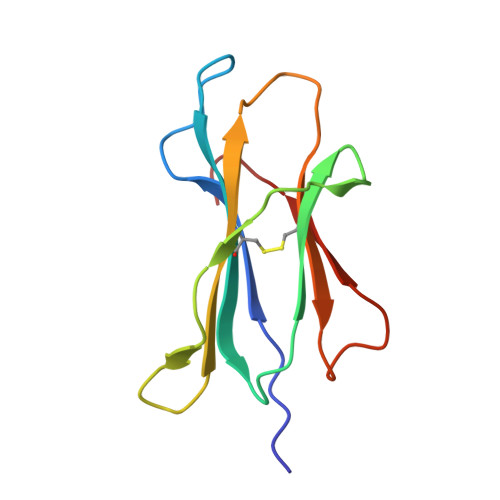
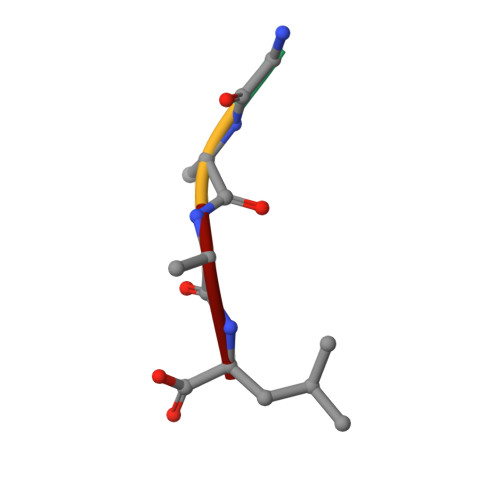
Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
Asa, M., Morita, D., Kuroha, J., Mizutani, T., Mori, N., Mikami, B., Sugita, M.(2022) J Biol Chem 298: 102100-102100
- PubMed: 35667438
- DOI: https://doi.org/10.1016/j.jbc.2022.102100
- Primary Citation of Related Structures:
7WJ2, 7WJ3, 7WT3, 7WT4, 7WT5 - PubMed Abstract:
Rhesus monkeys have evolved MHC-encoded class I allomorphs such as Mamu-B∗098 that are capable of binding N-myristoylated short lipopeptides rather than conventional long peptides; however, it remains unknown whether such antigen-binding molecules exist in other species, including humans. We herein demonstrate that human leukocyte antigen (HLA)-A∗24:02 and HLA-C∗14:02 proteins, which are known to bind conventional long peptides, also have the potential to bind N-myristoylated short lipopeptides. These HLA class I molecules shared a serine at position 9 (Ser9) with Mamu-B∗098, in contrast to most MHC class I molecules that harbor a larger amino acid residue, such as tyrosine, at this position. High resolution X-ray crystallographic analyses of lipopeptide-bound HLA-A∗24:02 and HLA-C∗14:02 complexes indicated that Ser9 was at the bottom of the B pocket with its small hydroxymethyl side chain directed away from the B-pocket cavity, thereby contributing to the formation of a deep hydrophobic cavity suitable for accommodating the long-chain fatty acid moiety of lipopeptide ligands. Upon peptide binding, however, we found the hydrogen-bond network involving Ser9 was reorganized, and the remodeled B pocket was able to capture the second amino acid residue (P2) of peptide ligands. Apart from the B pocket, virtually no marked alterations were observed for the A and F pockets upon peptide and lipopeptide binding. Thus, we concluded that the structural flexibility of the large B pocket of HLA-A∗2402 and HLA-C∗1402 primarily accounted for their previously unrecognized capacity to bind such chemically distinct ligands as conventional peptides and N-myristoylated lipopeptides.
Organizational Affiliation:
Laboratory of Cell Regulation, Institute for Life and Medical Sciences, Kyoto University, Kyoto, Japan; Laboratory of Cell Regulation and Molecular Network, Graduate School of Biostudies, Kyoto University, Kyoto, Japan.