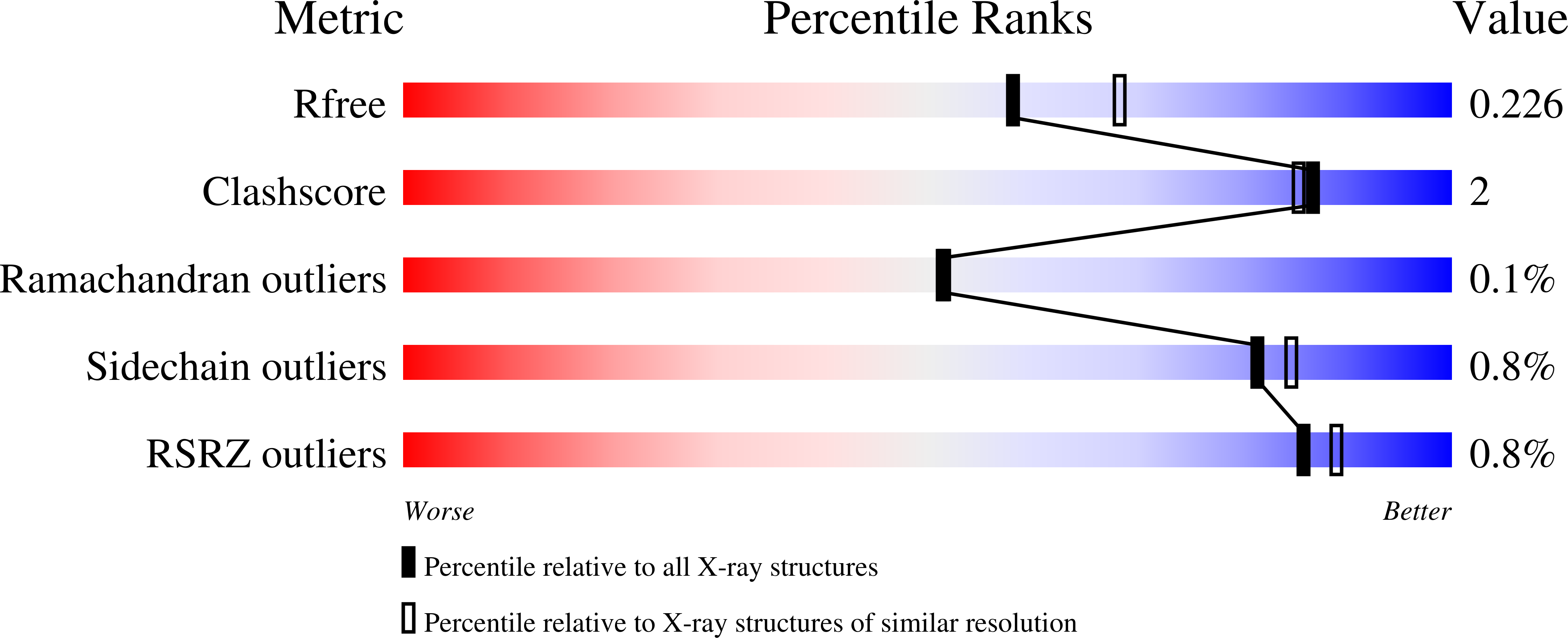
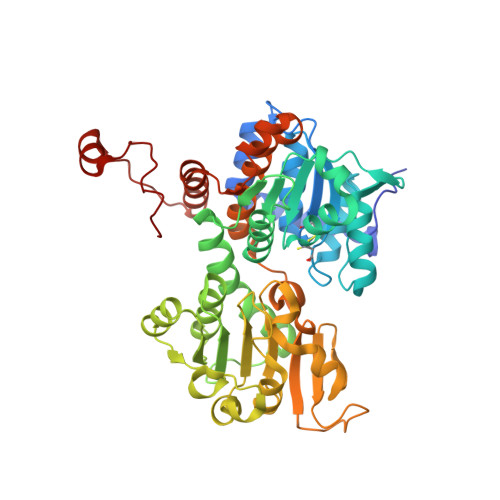
A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Gawel, M., Malecki, P.H., Sliwiak, J., Stepniewska, M., Imiolczyk, B., Czyrko-Horczak, J., Jakubczyk, D., Marczak, L., Plonska-Brzezinska, M.E., Brzezinski, K.(2024) Chem Commun (Camb) 60: 11504-11507
- PubMed: 39230573
- DOI: https://doi.org/10.1039/d4cc03143a
- Primary Citation of Related Structures:
7ZD0, 7ZD1, 7ZD2, 7ZD3, 7ZD4 - PubMed Abstract:
We report biochemical and structural studies on inhibiting bacterial S -adenosyl-L-homocysteine hydrolase by transition metal cations. Our results revealed diverse molecular mechanisms of enzyme inactivation. Depending on the cation, the mechanism is based on arresting the enzyme in its closed, inactive conformation, disulfide bond formation within the active site or oxidation of the intermediate form of a cofactor.
Organizational Affiliation:
Institute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14, 61-704 Poznan, Poland. [email protected].