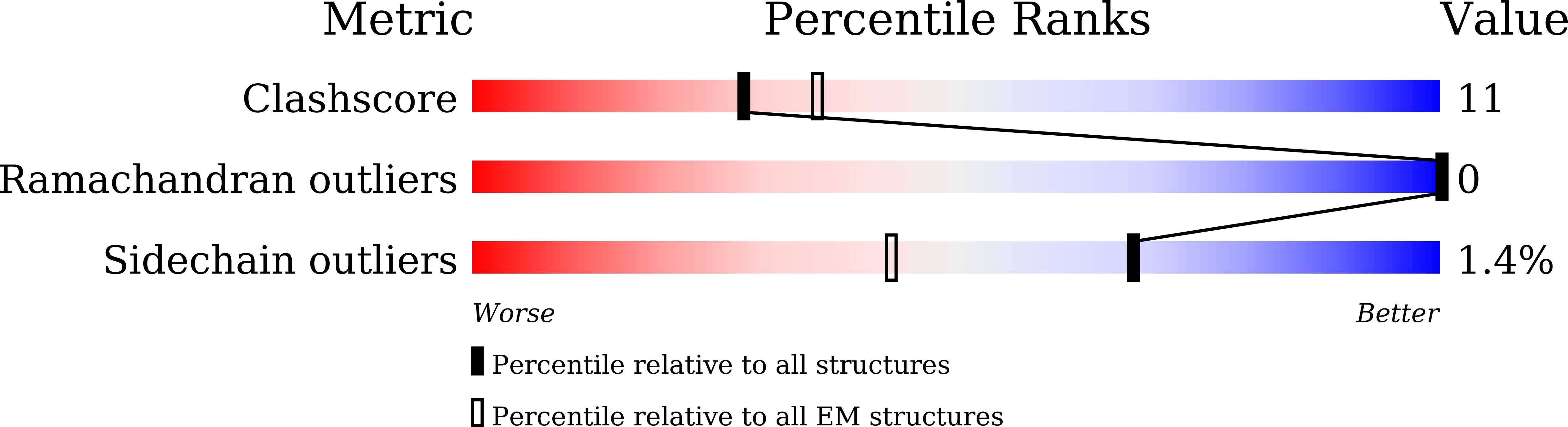A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex.
Tani, K., Nagashima, K.V.P., Kanno, R., Kawamura, S., Kikuchi, R., Hall, M., Yu, L.J., Kimura, Y., Madigan, M.T., Mizoguchi, A., Humbel, B.M., Wang-Otomo, Z.Y.(2021) Nat Commun 12: 6300-6300
- PubMed: 34728609
- DOI: https://doi.org/10.1038/s41467-021-26561-9
- Primary Citation of Related Structures:
7F0L - PubMed Abstract:
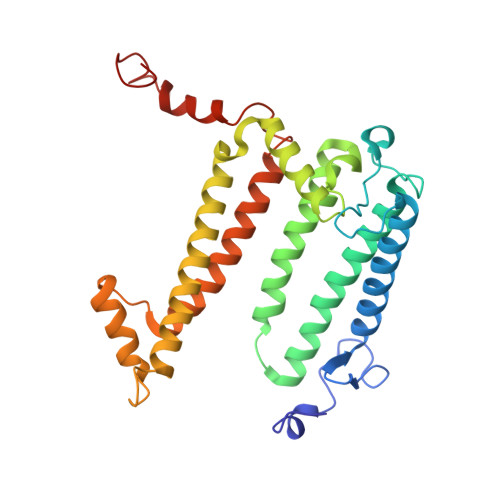
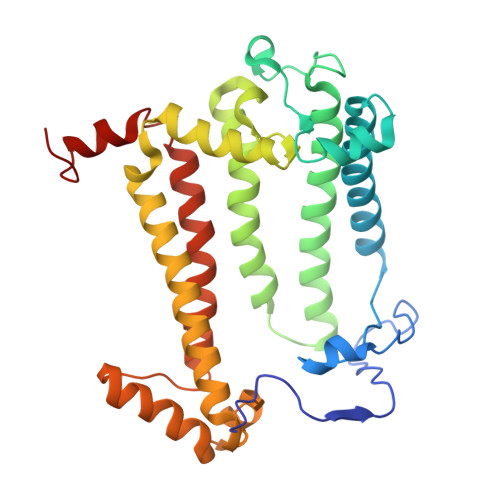
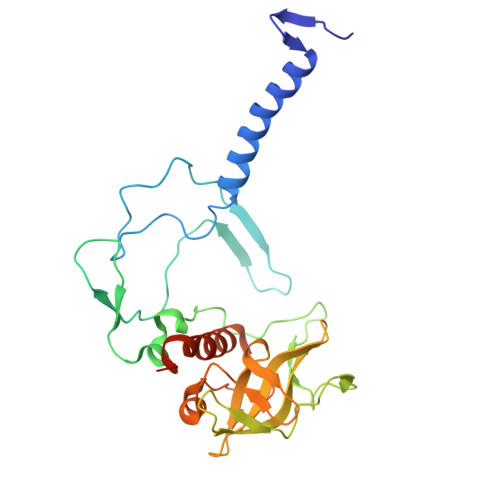
Rhodobacter (Rba.) sphaeroides is the most widely used model organism in bacterial photosynthesis. The light-harvesting-reaction center (LH1-RC) core complex of this purple phototroph is characterized by the co-existence of monomeric and dimeric forms, the presence of the protein PufX, and approximately two carotenoids per LH1 αβ-polypeptides. Despite many efforts, structures of the Rba. sphaeroides LH1-RC have not been obtained at high resolutions. Here we report a cryo-EM structure of the monomeric LH1-RC from Rba. sphaeroides strain IL106 at 2.9 Å resolution. The LH1 complex forms a C-shaped structure composed of 14 αβ-polypeptides around the RC with a large ring opening. From the cryo-EM density map, a previously unrecognized integral membrane protein, referred to as protein-U, was identified. Protein-U has a U-shaped conformation near the LH1-ring opening and was annotated as a hypothetical protein in the Rba. sphaeroides genome. Deletion of protein-U resulted in a mutant strain that expressed a much-reduced amount of the dimeric LH1-RC, indicating an important role for protein-U in dimerization of the LH1-RC complex. PufX was located opposite protein-U on the LH1-ring opening, and both its position and conformation differed from that of previous reports of dimeric LH1-RC structures obtained at low-resolution. Twenty-six molecules of the carotenoid spheroidene arranged in two distinct configurations were resolved in the Rba. sphaeroides LH1 and were positioned within the complex to block its channels. Our findings offer an exciting new view of the core photocomplex of Rba. sphaeroides and the connections between structure and function in bacterial photocomplexes in general.
Organizational Affiliation:
Graduate School of Medicine, Mie University, Tsu, 514-8507, Japan. [email protected].