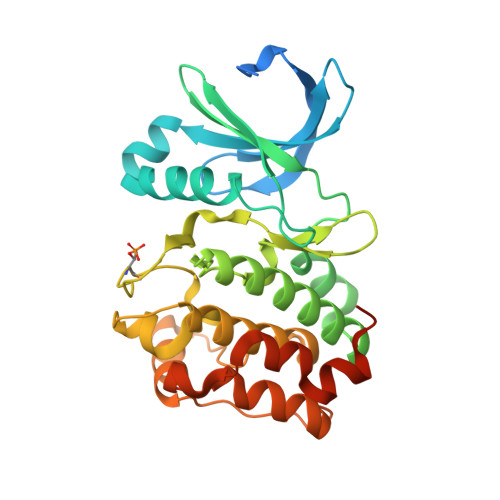
Crystal structure of a covalently linked Aurora-A-MYCN complex.
Diebold, M., Schonemann, L., Eilers, M., Sotriffer, C., Schindelin, H.(2023) Acta Crystallogr D Struct Biol 79: 1-9
- PubMed: 36601802
- DOI: https://doi.org/10.1107/S2059798322011433
- Primary Citation of Related Structures:
7ZTL - PubMed Abstract:
Formation of the Aurora-A-MYCN complex increases levels of the oncogenic transcription factor MYCN in neuroblastoma cells by abrogating its degradation through the ubiquitin proteasome system. While some small-molecule inhibitors of Aurora-A were shown to destabilize MYCN, clinical trials have not been satisfactory to date. MYCN itself is considered to be `undruggable' due to its large intrinsically disordered regions. Targeting the Aurora-A-MYCN complex rather than Aurora-A or MYCN alone will open new possibilities for drug development and screening campaigns. To overcome the challenges that a ternary system composed of Aurora-A, MYCN and a small molecule entails, a covalently cross-linked construct of the Aurora-A-MYCN complex was designed, expressed and characterized, thus enabling screening and design campaigns to identify selective binders.
Organizational Affiliation:
Institute of Pharmacy and Food Chemistry, University of Würzburg, Am Hubland, 97074 Würzburg, Germany.