Cholera toxin variants
Heim, J.B., Serrano, A., Kersten, F., Mojica, N., Cordara, G., Tatulian, S.A., Krengel, U., Teter, K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cholera enterotoxin subunit A | A, G [auth B] | 240 | Vibrio cholerae O1 | Mutation(s): 4 Gene Names: ctxA, toxA, VC_1457 | 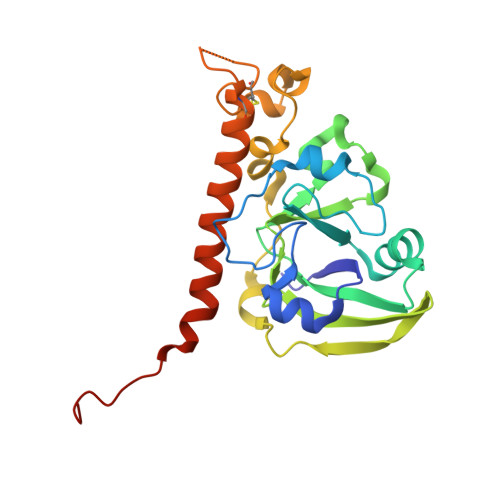 |
UniProt | |||||
Find proteins for P01555 (Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)) Explore P01555 Go to UniProtKB: P01555 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01555 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cholera enterotoxin subunit B | 103 | Vibrio cholerae O1 | Mutation(s): 2 Gene Names: ctxB, toxB, VC_1456 | 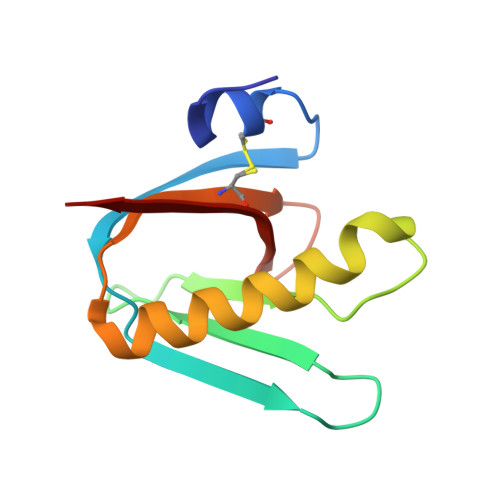 | |
UniProt | |||||
Find proteins for P01556 (Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)) Explore P01556 Go to UniProtKB: P01556 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01556 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | AA [auth B], DA [auth I], Q [auth E], W [auth H] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| GLA Query on GLA | BA [auth C] FA [auth K] HA [auth L] O [auth D] R [auth E] | alpha-D-galactopyranose C6 H12 O6 WQZGKKKJIJFFOK-PHYPRBDBSA-N |  | ||
| GAL Query on GAL | CA [auth C] EA [auth I] GA [auth K] IA [auth L] N [auth A] | beta-D-galactopyranose C6 H12 O6 WQZGKKKJIJFFOK-FPRJBGLDSA-N |  | ||
| NA Query on NA | M [auth A], Z [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.295 | α = 90 |
| b = 89.61 | β = 98.186 |
| c = 141.094 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Coot | model building |
| MxCuBE | data collection |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | R03AI112854 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | R01AI137056 |